RNTuple: Where are we now and what's next?
(26 January 2024)Hello, this is Florine from the ROOT team! Over the past year, I’ve been working as a technical student funded by ATLAS to evaluate and help further develop RNTuple. As you may already be aware, RNTuple [1] is currently being developed as the successor to TTree, and is projected to be used in Run 4. I imagine you might be wondering why there is a need for a completely new (TTree-incompatible) system, and what this looks like. That’s why in this blog post, I will try to answer this question, as well as give you an overview of the current status of RNTuple, what we’re still working on before its first production release (and what we will work on beyond this), and finally how you can already try it out!
Why do we need RNTuple?
At this point, ROOT has been around for more than a quarter of a century – and TTree for just as long. And as you might imagine, the computing landscape today looks vastly different compared to 25 years ago. Just to set the scene: when ROOT was first released, there was no C++ standard yet and parallel (let alone distributed) computing really wasn’t a thing yet. On the hardware side, modern storage technologies such as SSDs and object stores were still unheard of, and let’s not forget to mention the evolution of networking technologies! Naturally, TTree wasn’t designed and implemented with these things in mind. Now of course, over the years a lot of effort has been put into improving the performance and stability of TTree to make it compatible with modern computing practices as much as possible. However, there are limits to what is possible in this regard, especially given the fact that backwards- and forwards-compatibility are two major requirements for ROOT’s I/O system. This has led to the fact that with the High-Luminosity LHC on the horizon, where 90% of the total amount of LHC data is expected to be produced [2], we need to think about more optimized ways to store physics data. The challenge here is that this data is unique in the sense that events (or, in computer science terms, “entry” or “row”) are statistically independent of each other. At the same time one event typically contains many (complex) data structures, of which we often only need a small subset at a time, and we found out that standard technologies are not well-tuned for this type of data storage [3]. That is why we decided to combine the years of experience with TTree and various industry best-practices and invest in the next generation of high-energy physics data storage. Enter RNTuple!
Where we are now?
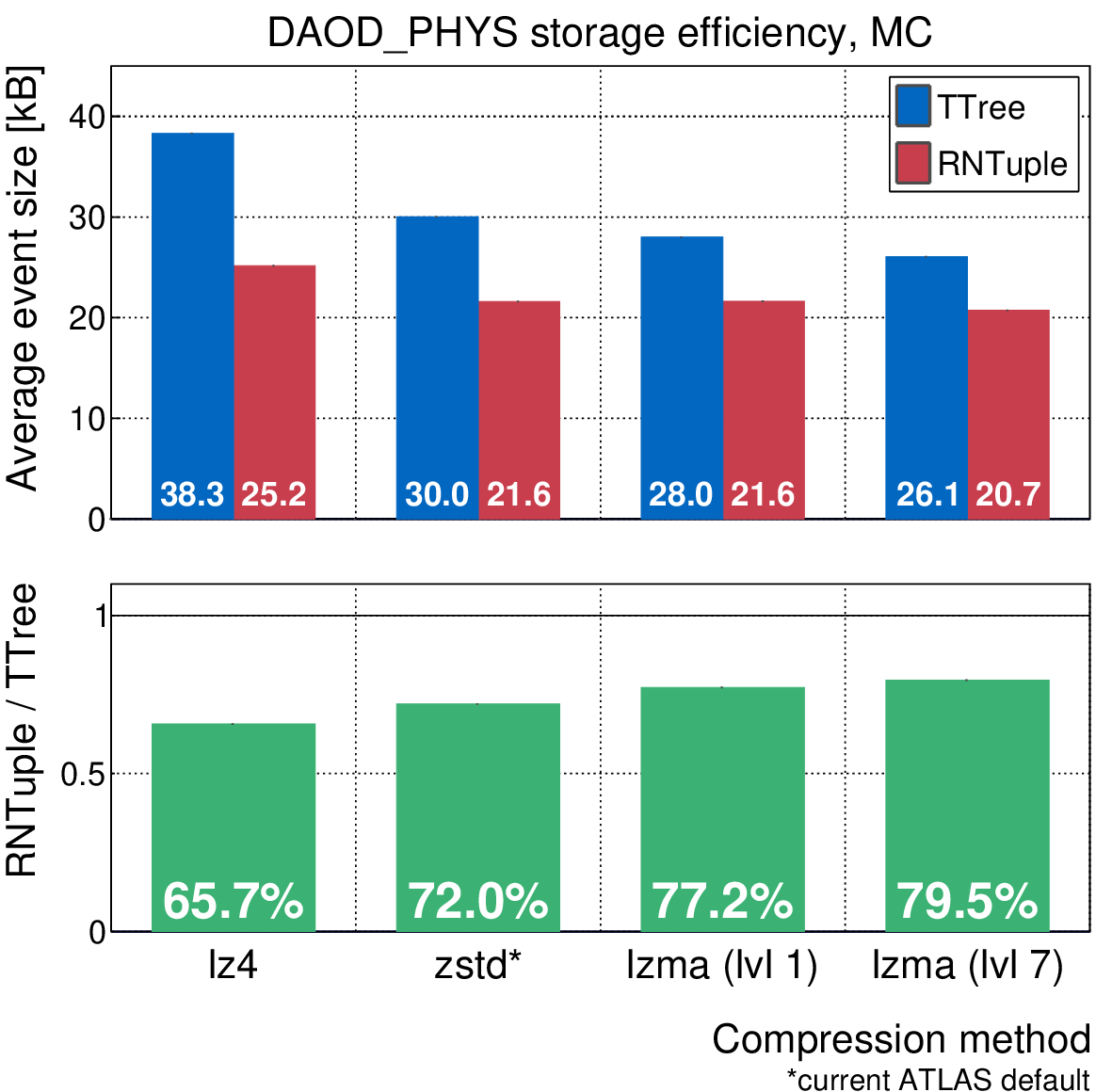
For the past four years, a lot of effort has been put into making RNTuple the best it can be. We are working closely with the experiments to make sure that RNTuple can support their data models across all relevant stages in the production pipeline. Simultaneously, we want to make sure that it is as optimized as possible. This means making sure that the data stored in RNTuple is as compact as possible, and at the same time coming up with ways in which we can make reading and writing RNTuples to and from memory as fast as possible. To give you an idea of where we’re currently at, the plot below shows the average on-disk event size for ATLAS’s DAOD_PHYS data model [4], comparing TTree and RNTuple. With RNTuple, we could potentially save 20-35% of storage space, and in turn reduce the consumed network bandwidth when reading the data from a remote location. When we’re talking about exabytes of event data, this is quite significant!
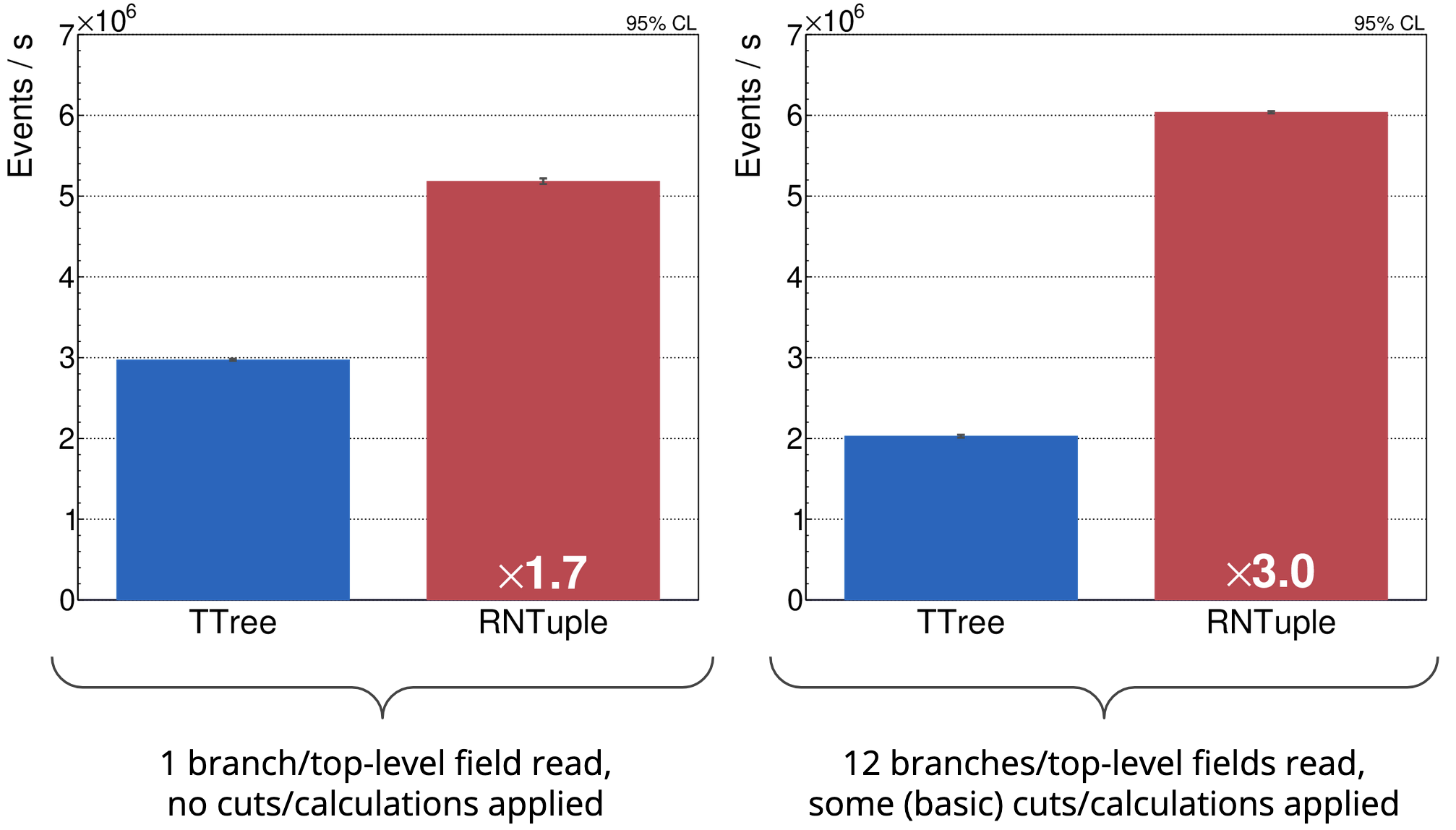
Besides storage efficiency, we’re also seeing very promising results when it comes to read throughput. The two plots below show the number of events processed per second for two different types of tasks, comparing ATLAS DAOD_PHYSLITE data sets stored in TTree and RNTuple (stored on an SSD). As you can see, RNTuple is remarkably faster than TTree, and similar observations are made for other data sets [1], [5].
Beyond performance, we have also been working hard on RNTuple’s interface and supported features. This includes compatibility with RDataFrame, being able to read and write C++ STL types as well as user-defined types and various other features to support existing experiment frameworks.
Can I try it out?
Yes! To be able to read and write RNTuples, the first thing you’ll need is a ROOT
installation that includes the
ROOT 7 experimental features enabled.
This is the case for the default LXPLUS installation, which runs ROOT’s (at the
time of writing) latest release, 6.30.02!
If you are running ROOT in a different way, you can easily check if ROOT 7 is
enabled for your installation by running root-config --has-root7 in your terminal.
If this returns yes, you’re all set! If you get a no, you will need to use a different
installation of ROOT that does. Check out the ROOT installation page
to get it. We strongly recommend using the most recent release in order to get
the latest and greatest from RNTuple.
Now, on to the fun part: using RNTuple! Of course, you could write a new RNTuple
completely from scratch, using fields and data that you come up with. This is
done using the RNTupleWriter
interface. Reading an RNTuple is then naturally done through the
RNTupleReader.
To get an idea of what this looks like in practice, check out for example
this tutorial.
Of course, it would be more interesting to try out RNTuple with real data, for
example with data from an analysis ntuple that is currently stored as a TTree.
Well, good news! RNTuple also comes with an RNTupleImporter
class that allows you to automatically convert your TTrees to RNTuples. This
can be as simple as executing the following two lines in the ROOT prompt. The
input file containing the source TTree is read remotely, meaning you can
directly copy-paste these lines into your ROOT prompt. Of course, it’s entirely
possible to use your own existing TTrees.
root [0] auto importer = ROOT::Experimental::RNTupleImporter::Create(
"http://root.cern/files/HiggsTauTauReduced/GluGluToHToTauTau.root",
"Events",
"my_rntuple.root")
root [1] importer->Import()
This will convert your TTree (called Events here) into an RNTuple also called
Events and write it to my_rntuple.root. Easy enough, but maybe you want more
control over this newly created RNTuple. For example, you might want to change
its name, or set the compression settings to something other than the default.
This (and more) can all be tweaked! Check out
the reference
or this tutorial to see
what options are possible.
Now, I already mentioned that we have been working on RNTuple compatibility with RDataFrame. Currently, with just one line change, you will be able to use your existing analysis code with data stored in RNTuple:
// Change this:
ROOT::RDataFrame df("Events", "http://root.cern/files/HiggsTauTauReduced/GluGluToHToTauTau.root");
// To this to use the RNTuple you just imported into "my_rntuple.root":
ROOT::RDataFrame df = ROOT::RDF::Experimental::FromRNTuple("Events", "my_rntuple.root");
// Use your existing analysis as-is!
💡 The automatic detection of RNTuples in RDataFrame is currently available in ROOT’s
masterbranch and will be available in ROOT 6.32.00!
Next steps for RNTuple
So, what’s next? Performance is always one of our main concerns. We are currently working on parallelizing the writing of RNTuples. In addition, we are working on what we like to call “interface ergonomics”, i.e. the way developers will interact with RNTuple. Be aware that this means that the RNTuple interfaces might still change a little in the coming months! Next to all of this, we are preparing for larger-scale performance testing to see in what areas we could further improve. Another area of work for the near future will be in the direction of data set combinatorics – that is, finding smart(er) ways of accessing and combining existing RNTuple data. And of course, we will continue to work with the experiments to make sure the transition to RNTuple will be as smooth as possible.
To wrap things up, things are looking good for RNTuple, and while there is still enough work to be done, we’re excited and eager to make RNTuple as good as it can be! If you want to know more about the evolution and performance of RNTuple, be sure to check out the references below, as well as our other publications. If you are eager to dive deeper into the specifics of the RNTuple binary format, you can read the specification here. Finally, reach out to us on the forum if you have any questions or if you would like to contribute to RNTuple or ROOT in general!
References
[1] J. Blomer, P. Canal, A. Naumann, and D. Piparo, “Evolution of the ROOT Tree I/O,” EPJ Web Conf., vol. 245, 2020, doi: 10.1051/epjconf/202024502030.
[2] ATLAS Collaboration, “ATLAS Software and Computing HL-LHC Roadmap,” CERN, Geneva, CERN-LHCC-2022-005, LHCC-G-182, 2022. Accessed: May 02, 2023. [Online]. Available: http://cds.cern.ch/record/2802918.
[3] J. Blomer, “A quantitative review of data formats for HEP analyses,” J. Phys. Conf. Ser., vol. 1085, p. 032020, Sep. 2018, doi: 10.1088/1742-6596/1085/3/032020.
[4] J. Elmsheuser et al., “Evolution of the ATLAS analysis model for Run-3 and prospects for HL-LHC,” EPJ Web Conf., vol. 245, 2020, doi: 10.1051/epjconf/202024506014.
[5] J. Lopez-Gomez and J. Blomer, “RNTuple performance: Status and Outlook.” arXiv, Apr. 07, 2022. doi: 10.48550/arXiv.2204.09043.
