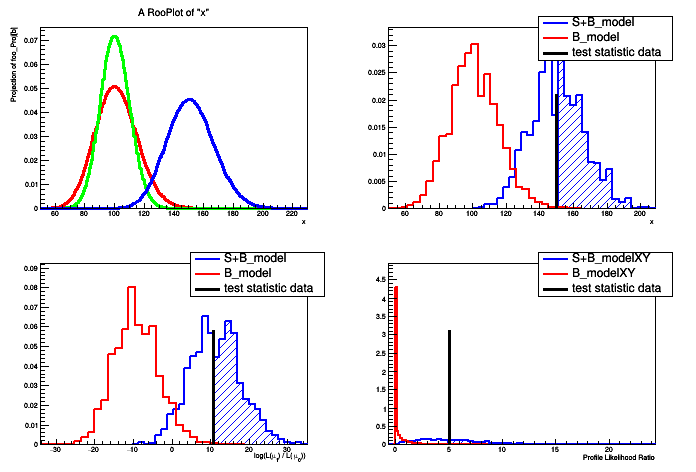
A hypothesis testing example based on number counting with background uncertainty.
NOTE: This example must be run with the ACLIC (the + option ) due to the new class that is defined.
The basic setup here is that a main measurement has observed x events with an expectation of s+b. One can choose an ad hoc prior for the uncertainty on b, or try to base it on an auxiliary measurement. In this case, the auxiliary measurement (aka control measurement, sideband) is another counting experiment with measurement y and expectation tau*b. With an 'original prior' on b, called \(\eta(b)\) then one can obtain a posterior from the auxiliary measurement \(\pi(b) = \eta(b) * Pois(y|tau*b).\) This is a principled choice for a prior on b in the main measurement of x, which can then be treated in a hybrid Bayesian/Frequentist way. Additionally, one can try to treat the two measurements simultaneously, which is detailed in Part 6 of the tutorial.
This tutorial is related to the FourBin.C tutorial in the modeling, but focuses on hypothesis testing instead of interval estimation.
More background on this 'prototype problem' can be found in the following papers:
Processing /mnt/build/workspace/root-makedoc-v612/rootspi/rdoc/src/v6-12-00-patches/tutorials/roostats/HybridInstructional.C...
�[1mRooFit v3.60 -- Developed by Wouter Verkerke and David Kirkby�[0m
Copyright (C) 2000-2013 NIKHEF, University of California & Stanford University
All rights reserved, please read http://roofit.sourceforge.net/license.txt
-----------------------------------------
Part 2
Hybrid p-value from direct integration = 0.00094165
Z_Gamma Significance = 3.10804
-----------------------------------------
Part 3
Z_Bi p-value (analytic): 0.00094165
Z_Bi significance (analytic): 3.10804
Real time 0:00:06, CP time 6.190
[#0] WARNING:Eval -- RooStatsUtils::MakeNuisancePdf - no constraints found on nuisance parameters in the input model
[#0] WARNING:Eval -- RooStatsUtils::MakeNuisancePdf - no constraints found on nuisance parameters in the input model
-----------------------------------------
Part 4
Results HypoTestCalculator_result:
- Null p-value = 0.00115 +/- 0.000239654
- Significance = 3.04848 +/- 0.0626148 sigma
- Number of Alt toys: 1000
- Number of Null toys: 20000
- Test statistic evaluated on data: 150
- CL_b: 0.00115 +/- 0.000239654
- CL_s+b: 0.524 +/- 0.0157932
- CL_s: 455.652 +/- 95.9434
Real time 0:00:03, CP time 3.130
[#0] WARNING:Eval -- RooStatsUtils::MakeNuisancePdf - no constraints found on nuisance parameters in the input model
[#0] WARNING:Eval -- RooStatsUtils::MakeNuisancePdf - no constraints found on nuisance parameters in the input model
-----------------------------------------
Part 5
Results HypoTestCalculator_result:
- Null p-value = 0.0009 +/- 0.000212037
- Significance = 3.12139 +/- 0.0693716 sigma
- Number of Alt toys: 1000
- Number of Null toys: 20000
- Test statistic evaluated on data: 10.8198
- CL_b: 0.0009 +/- 0.000212037
- CL_s+b: 0.535 +/- 0.0157726
- CL_s: 594.444 +/- 141.141
Real time 0:00:03, CP time 3.620
[#0] WARNING:Eval -- RooStatsUtils::MakeNuisancePdf - no constraints found on nuisance parameters in the input model
[#0] WARNING:Eval -- RooStatsUtils::MakeNuisancePdf - no constraints found on nuisance parameters in the input model
-----------------------------------------
Part 6
Results HypoTestCalculator_result:
- Null p-value = 0.000666667 +/- 0.000149021
- Significance = 3.20871 +/- 0.0642752 sigma
- Number of Alt toys: 1000
- Number of Null toys: 30000
- Test statistic evaluated on data: 5.03388
- CL_b: 0.000666667 +/- 0.000149021
- CL_s+b: 0.489 +/- 0.0158076
- CL_s: 733.5 +/- 165.667
Real time 0:00:35, CP time 35.260
public:
BinCountTestStat(void) : fColumnName("tmp") {}
BinCountTestStat(string columnName) : fColumnName(columnName) {}
}
return value;
}
virtual const TString GetVarName() const { return fColumnName; }
private:
string fColumnName;
protected:
};
void HybridInstructional() {
w->
factory(
"Poisson::px(x[150,0,500],sum::splusb(s[0,0,100],b[100,0,300]))");
w->
factory(
"Poisson::py(y[100,0,500],prod::taub(tau[1.],b))");
w->
factory(
"PROJ::averagedModel(PROD::foo(px|b,py,prior_b),b)") ;
cout << "-----------------------------------------"<<endl;
cout << "Part 2" << endl;
cout <<
"Hybrid p-value from direct integration = " << 1-cdf->
getVal() << endl;
cout << "Z_Gamma Significance = " <<
cout << "-----------------------------------------"<<endl;
cout << "Part 3" << endl;
std::cout << "Z_Bi p-value (analytic): " << p_Bi << std::endl;
std::cout << "Z_Bi significance (analytic): " << Z_Bi << std::endl;
b_model.SetPdf(*w->
pdf(
"px"));
b_model.SetObservables(*w->
set(
"obs"));
b_model.SetParametersOfInterest(*w->
set(
"poi"));
b_model.SetSnapshot(*w->
set(
"poi"));
sb_model.SetPdf(*w->
pdf(
"px"));
sb_model.SetObservables(*w->
set(
"obs"));
sb_model.SetParametersOfInterest(*w->
set(
"poi"));
sb_model.SetSnapshot(*w->
set(
"poi"));
BinCountTestStat binCount("x");
w->
factory(
"Gaussian::gauss_prior(b,y, expr::sqrty('sqrt(y)',y))");
w->
factory(
"Lognormal::lognorm_prior(b,y, expr::kappa('1+1./sqrt(y)',y))");
hc1.SetToys(20000,1000);
hc1.ForcePriorNuisanceAlt(*w->
pdf(
"py"));
hc1.ForcePriorNuisanceNull(*w->
pdf(
"py"));
cout << "-----------------------------------------"<<endl;
cout << "Part 4" << endl;
slrts.SetAltParameters(*sb_model.GetSnapshot());
hc2.SetToys(20000,1000);
hc2.ForcePriorNuisanceAlt(*w->
pdf(
"py"));
hc2.ForcePriorNuisanceNull(*w->
pdf(
"py"));
cout << "-----------------------------------------"<<endl;
cout << "Part 5" << endl;
dataXY->
add(*w->
set(
"obsXY"));
b_modelXY.SetPdf(*w->
pdf(
"model"));
b_modelXY.SetObservables(*w->
set(
"obsXY"));
b_modelXY.SetParametersOfInterest(*w->
set(
"poi"));
b_modelXY.SetSnapshot(*w->
set(
"poi"));
sb_modelXY.SetPdf(*w->
pdf(
"model"));
sb_modelXY.SetObservables(*w->
set(
"obsXY"));
sb_modelXY.SetParametersOfInterest(*w->
set(
"poi"));
sb_modelXY.SetSnapshot(*w->
set(
"poi"));
ropl(*b_modelXY.GetPdf(), *sb_modelXY.GetPdf(), sb_modelXY.GetSnapshot());
w->
factory(
"Gamma::gamma_y0(b,sum::temp0(y0,1),1,0)");
w->
factory(
"Gaussian::gauss_prior_y0(b,y0, expr::sqrty0('sqrt(y0)',y0))");
hc3.SetToys(30000,1000);
hc3.ForcePriorNuisanceAlt(*w->
pdf(
"gamma_y0"));
hc3.ForcePriorNuisanceNull(*w->
pdf(
"gamma_y0"));
cout << "-----------------------------------------"<<endl;
cout << "Part 6" << endl;
}



 Example demonstrating usage of HybridCalcultor
Example demonstrating usage of HybridCalcultor