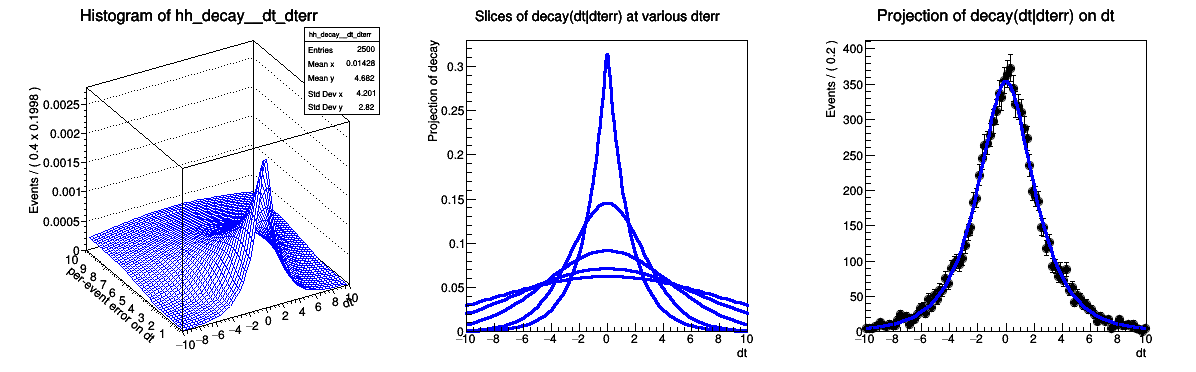
Complete example with use of conditional p.d.f. with per-event errors
Processing /mnt/build/workspace/root-makedoc-v612/rootspi/rdoc/src/v6-12-00-patches/tutorials/roofit/rf306_condpereventerrors.C...
�[1mRooFit v3.60 -- Developed by Wouter Verkerke and David Kirkby�[0m
Copyright (C) 2000-2013 NIKHEF, University of California & Stanford University
All rights reserved, please read http://roofit.sourceforge.net/license.txt
[#1] INFO:Minization -- RooMinimizer::optimizeConst: activating const optimization
**********
** 1 **SET PRINT 1
**********
**********
** 2 **SET NOGRAD
**********
PARAMETER DEFINITIONS:
NO. NAME VALUE STEP SIZE LIMITS
1 bias 0.00000e+00 2.00000e+00 -1.00000e+01 1.00000e+01
2 sigma 1.00000e+00 4.50000e-01 1.00000e-01 1.00000e+01
**********
** 3 **SET ERR 0.5
**********
**********
** 4 **SET PRINT 1
**********
**********
** 5 **SET STR 1
**********
NOW USING STRATEGY 1: TRY TO BALANCE SPEED AGAINST RELIABILITY
**********
** 6 **MIGRAD 1000 1
**********
FIRST CALL TO USER FUNCTION AT NEW START POINT, WITH IFLAG=4.
START MIGRAD MINIMIZATION. STRATEGY 1. CONVERGENCE WHEN EDM .LT. 1.00e-03
FCN=23876.4 FROM MIGRAD STATUS=INITIATE 8 CALLS 9 TOTAL
EDM= unknown STRATEGY= 1 NO ERROR MATRIX
EXT PARAMETER CURRENT GUESS STEP FIRST
NO. NAME VALUE ERROR SIZE DERIVATIVE
1 bias 0.00000e+00 2.00000e+00 2.01358e-01 -1.70342e+02
2 sigma 1.00000e+00 4.50000e-01 1.63378e-01 8.62474e+01
ERR DEF= 0.5
MIGRAD MINIMIZATION HAS CONVERGED.
MIGRAD WILL VERIFY CONVERGENCE AND ERROR MATRIX.
COVARIANCE MATRIX CALCULATED SUCCESSFULLY
FCN=23876.2 FROM MIGRAD STATUS=CONVERGED 30 CALLS 31 TOTAL
EDM=3.03467e-08 STRATEGY= 1 ERROR MATRIX ACCURATE
EXT PARAMETER STEP FIRST
NO. NAME VALUE ERROR SIZE DERIVATIVE
1 bias 5.26463e-03 1.72103e-02 1.83632e-04 1.00797e-01
2 sigma 9.87130e-01 2.04183e-02 7.70435e-04 2.52620e-03
ERR DEF= 0.5
EXTERNAL ERROR MATRIX. NDIM= 25 NPAR= 2 ERR DEF=0.5
2.962e-04 -4.402e-06
-4.402e-06 4.169e-04
PARAMETER CORRELATION COEFFICIENTS
NO. GLOBAL 1 2
1 0.01253 1.000 -0.013
2 0.01253 -0.013 1.000
**********
** 7 **SET ERR 0.5
**********
**********
** 8 **SET PRINT 1
**********
**********
** 9 **HESSE 1000
**********
COVARIANCE MATRIX CALCULATED SUCCESSFULLY
FCN=23876.2 FROM HESSE STATUS=OK 12 CALLS 43 TOTAL
EDM=3.12688e-08 STRATEGY= 1 ERROR MATRIX ACCURATE
EXT PARAMETER INTERNAL INTERNAL
NO. NAME VALUE ERROR STEP SIZE VALUE
1 bias 5.26463e-03 1.72105e-02 3.67263e-05 5.26463e-04
2 sigma 9.87130e-01 2.04260e-02 3.08174e-05 -9.62778e-01
ERR DEF= 0.5
EXTERNAL ERROR MATRIX. NDIM= 25 NPAR= 2 ERR DEF=0.5
2.962e-04 -4.786e-06
-4.786e-06 4.172e-04
PARAMETER CORRELATION COEFFICIENTS
NO. GLOBAL 1 2
1 0.01361 1.000 -0.014
2 0.01361 -0.014 1.000
[#1] INFO:Minization -- RooMinimizer::optimizeConst: deactivating const optimization
[#1] INFO:NumericIntegration -- RooRealIntegral::init(gm1_conv_exp(-abs(@0)/@1)_dt_tau_[decay_gm]_Int[dt,dterr]) using numeric integrator RooIntegrator1D to calculate Int(dterr)
[#1] INFO:Plotting -- RooAbsReal::plotOn(decay_gm) plot on dt averages using data variables (dterr)
[#1] INFO:Plotting -- RooDataWeightedAverage::ctor(decay_gmDataWgtAvg) constructing data weighted average of function decay_gm_Norm[dt] over 100 data points of (dterr) with a total weight of 10000
.........................................................................................................................................................................................................................



 'MULTIDIMENSIONAL MODELS' RooFit tutorial macro #306
'MULTIDIMENSIONAL MODELS' RooFit tutorial macro #306