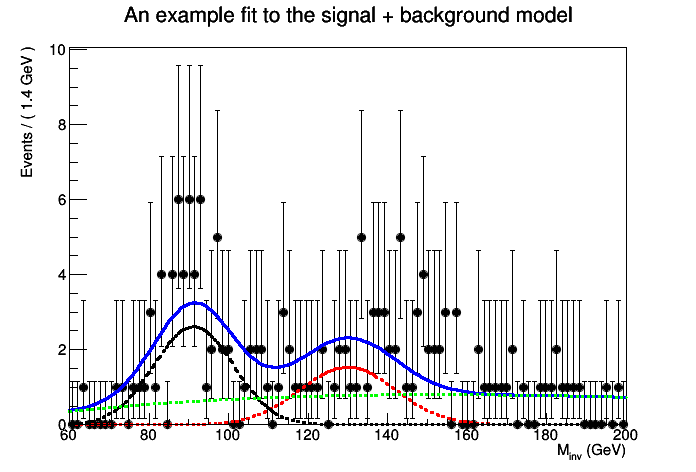
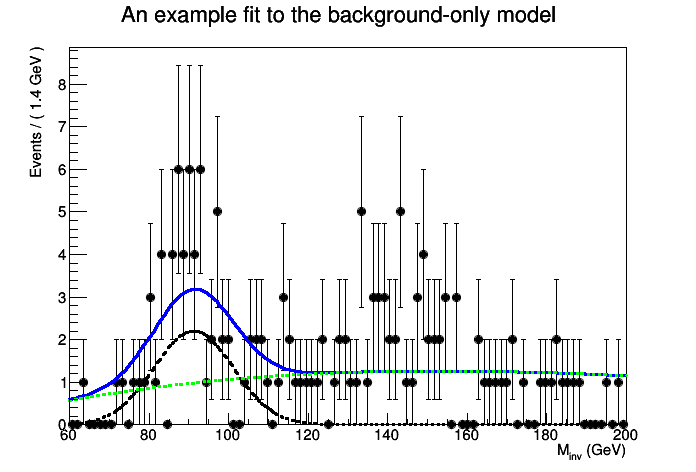
This tutorial macro shows a typical search for a new particle by studying an invariant mass distribution. The macro creates a simple signal model and two background models, which are added to a RooWorkspace. The macro creates a toy dataset, and then uses a RooStats ProfileLikleihoodCalculator to do a hypothesis test of the background-only and signal+background hypotheses. In this example, shape uncertainties are not taken into account, but normalization uncertainties are.
Processing /mnt/build/workspace/root-makedoc-v612/rootspi/rdoc/src/v6-12-00-patches/tutorials/roostats/rs102_hypotestwithshapes.C...
�[1mRooFit v3.60 -- Developed by Wouter Verkerke and David Kirkby�[0m
Copyright (C) 2000-2013 NIKHEF, University of California & Stanford University
All rights reserved, please read http://roofit.sourceforge.net/license.txt
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooAddPdf::model
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooGaussian::sigModel
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooRealVar::invMass
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooRealVar::mH
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooRealVar::sigma1
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooProduct::fsig
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooRealVar::mu
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooRealVar::ratioSigEff
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooRealVar::fsigExpected
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooGaussian::zjjModel
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooRealVar::mZ
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooRealVar::sigma1_z
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooRealVar::fzjj
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooChebychev::qcdModel
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooRealVar::a0
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooRealVar::a1
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooRealVar::a2
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing dataset modelData
[#1] INFO:ObjectHandling -- RooWorkSpace::import(myWS) changing name of dataset from modelData to data
[#1] INFO:Minization -- createNLL: caching constraint set under name CONSTR_OF_PDF_model_FOR_OBS_invMass with 0 entries
[#0] PROGRESS:Minization -- ProfileLikelihoodCalcultor::DoGLobalFit - find MLE
[#0] PROGRESS:Minization -- ProfileLikelihoodCalcultor::DoMinimizeNLL - using Minuit / Migrad with strategy 1
[#1] INFO:Minization -- RooMinimizer::optimizeConst: activating const optimization
[#1] INFO:Minization -- The following expressions have been identified as constant and will be precalculated and cached: (sigModel,zjjModel,qcdModel)
[#1] INFO:Minization --
RooFitResult: minimized FCN value: 717.039, estimated distance to minimum: 8.90226e-10
covariance matrix quality: Full, accurate covariance matrix
Status : MINIMIZE=0
Floating Parameter FinalValue +/- Error
-------------------- --------------------------
fzjj 3.1152e-01 +/- 5.03e-02
mu 1.0968e+00 +/- 3.03e-01
[#0] PROGRESS:Minization -- ProfileLikelihoodCalcultor::GetHypoTest - do conditional fit
[#0] PROGRESS:Minization -- ProfileLikelihoodCalcultor::DoMinimizeNLL - using Minuit / Migrad with strategy 1
[#1] INFO:Minization -- RooMinimizer::optimizeConst: activating const optimization
[#1] INFO:Minization --
RooFitResult: minimized FCN value: 723.97, estimated distance to minimum: 2.09863e-09
covariance matrix quality: Full, accurate covariance matrix
Status : MINIMIZE=0
Floating Parameter FinalValue +/- Error
-------------------- --------------------------
fzjj 2.6213e-01 +/- 5.18e-02
-------------------------------------------------
The p-value for the null is 9.83108e-05
Corresponding to a significance of 3.72332
-------------------------------------------------
[#1] INFO:Minization -- createNLL picked up cached consraints from workspace with 0 entries
[#1] INFO:Minization -- RooMinimizer::optimizeConst: activating const optimization
[#1] INFO:Minization -- The following expressions have been identified as constant and will be precalculated and cached: (sigModel,zjjModel,qcdModel)
[#1] INFO:Minization -- RooMinimizer::optimizeConst: deactivating const optimization
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model) directly selected PDF components: (sigModel)
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model) indirectly selected PDF components: ()
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model) directly selected PDF components: (zjjModel)
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model) indirectly selected PDF components: ()
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model) directly selected PDF components: (qcdModel)
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model) indirectly selected PDF components: ()
[#1] INFO:Minization -- createNLL picked up cached consraints from workspace with 0 entries
[#1] INFO:Minization -- RooMinimizer::optimizeConst: activating const optimization
[#1] INFO:Minization -- The following expressions have been identified as constant and will be precalculated and cached: (sigModel,zjjModel,qcdModel)
[#1] INFO:Minization -- RooMinimizer::optimizeConst: deactivating const optimization
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model) directly selected PDF components: (zjjModel)
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model) indirectly selected PDF components: ()
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model) directly selected PDF components: (qcdModel)
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model) indirectly selected PDF components: ()



 rs102_hypotestwithshapes for RooStats project
rs102_hypotestwithshapes for RooStats project