| rs101_limitexample.C: 'Limit Example' RooStats tutorial macro #101 | Roostats tutorials | rs301_splot.C: SPlot tutorial |
rs102_hypotestwithshapes.C: rs102_hypotestwithshapes for RooStats project
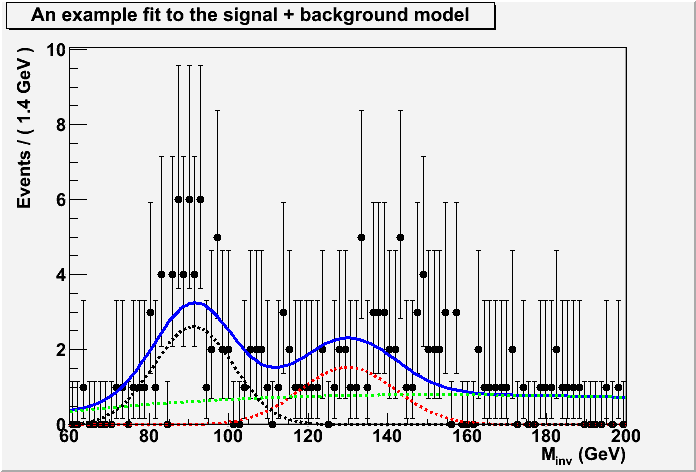
///////////////////////////////////////////////////////////////// // // rs102_hypotestwithshapes for RooStats project // Author: Kyle Cranmer <cranmer@cern.ch> // // Modified from version of February 29, 2008 // // This tutorial macro shows a typical search for a new particle // by studying an invariant mass distribution. // The macro creates a simple signal model and two background models, // which are added to a RooWorkspace. // The macro creates a toy dataset, and then uses a RooStats // ProfileLikleihoodCalculator to do a hypothesis test of the // background-only and signal+background hypotheses. // In this example, shape uncertainties are not taken into account, but // normalization uncertainties are. // ///////////////////////////////////////////////////////////////// #ifndef __CINT__ #include "RooGlobalFunc.h" #endif #include "RooDataSet.h" #include "RooRealVar.h" #include "RooGaussian.h" #include "RooAddPdf.h" #include "RooProdPdf.h" #include "RooAddition.h" #include "RooProduct.h" #include "TCanvas.h" #include "RooChebychev.h" #include "RooAbsPdf.h" #include "RooFit.h" #include "RooFitResult.h" #include "RooPlot.h" #include "RooAbsArg.h" #include "RooWorkspace.h" #include "RooStats/ProfileLikelihoodCalculator.h" #include "RooStats/HypoTestResult.h" #include <string> // use this order for safety on library loading using namespace RooFit; using namespace RooStats; // see below for implementation void AddModel(RooWorkspace*); void AddData(RooWorkspace*); void DoHypothesisTest(RooWorkspace*); void MakePlots(RooWorkspace*); //____________________________________ void rs102_hypotestwithshapes() { // The main macro. // Create a workspace to manage the project. RooWorkspace* wspace = new RooWorkspace("myWS"); // add the signal and background models to the workspace AddModel(wspace); // add some toy data to the workspace AddData(wspace); // inspect the workspace if you wish // wspace->Print(); // do the hypothesis test DoHypothesisTest(wspace); // make some plots MakePlots(wspace); // cleanup delete wspace; } //____________________________________ void AddModel(RooWorkspace* wks){ // Make models for signal (Higgs) and background (Z+jets and QCD) // In real life, this part requires an intellegent modeling // of signal and background -- this is only an example. // set range of observable Double_t lowRange = 60, highRange = 200; // make a RooRealVar for the observable RooRealVar invMass("invMass", "M_{inv}", lowRange, highRange,"GeV"); ///////////////////////////////////////////// // make a simple signal model. RooRealVar mH("mH","Higgs Mass",130,90,160) ; RooRealVar sigma1("sigma1","Width of Gaussian",12.,2,100) ; RooGaussian sigModel("sigModel", "Signal Model", invMass, mH, sigma1); // we will test this specific mass point for the signal mH.setConstant(); // and we assume we know the mass resolution sigma1.setConstant(); ///////////////////////////////////////////// // make zjj model. Just like signal model RooRealVar mZ("mZ", "Z Mass", 91.2, 0, 100); RooRealVar sigma1_z("sigma1_z","Width of Gaussian",10.,6,100) ; RooGaussian zjjModel("zjjModel", "Z+jets Model", invMass, mZ, sigma1_z); // we know Z mass mZ.setConstant(); // assume we know resolution too sigma1_z.setConstant(); ////////////////////////////////////////////// // make QCD model RooRealVar a0("a0","a0",0.26,-1,1) ; RooRealVar a1("a1","a1",-0.17596,-1,1) ; RooRealVar a2("a2","a2",0.018437,-1,1) ; RooRealVar a3("a3","a3",0.02,-1,1) ; RooChebychev qcdModel("qcdModel","A Polynomail for QCD",invMass,RooArgList(a0,a1,a2)) ; // let's assume this shape is known, but the normalization is not a0.setConstant(); a1.setConstant(); a2.setConstant(); ////////////////////////////////////////////// // combined model // Setting the fraction of Zjj to be 40% for initial guess. RooRealVar fzjj("fzjj","fraction of zjj background events",.4,0.,1) ; // Set the expected fraction of signal to 20%. RooRealVar fsigExpected("fsigExpected","expected fraction of signal events",.2,0.,1) ; fsigExpected.setConstant(); // use mu as main parameter, so fix this. // Introduce mu: the signal strength in units of the expectation. // eg. mu = 1 is the SM, mu = 0 is no signal, mu=2 is 2x the SM RooRealVar mu("mu","signal strength in units of SM expectation",1,0.,2) ; // Introduce ratio of signal efficiency to nominal signal efficiency. // This is useful if you want to do limits on cross section. RooRealVar ratioSigEff("ratioSigEff","ratio of signal efficiency to nominal signal efficiency",1. ,0.,2) ; ratioSigEff.setConstant(kTRUE); // finally the signal fraction is the product of the terms above. RooProduct fsig("fsig","fraction of signal events",RooArgSet(mu,ratioSigEff,fsigExpected)) ; // full model RooAddPdf model("model","sig+zjj+qcd background shapes",RooArgList(sigModel,zjjModel, qcdModel),RooArgList(fsig,fzjj)) ; // interesting for debugging and visualizing the model // model.printCompactTree("","fullModel.txt"); // model.graphVizTree("fullModel.dot"); wks->import(model); } //____________________________________ void AddData(RooWorkspace* wks){ // Add a toy dataset Int_t nEvents = 150; RooAbsPdf* model = wks->pdf("model"); RooRealVar* invMass = wks->var("invMass"); RooDataSet* data = model->generate(*invMass,nEvents); wks->import(*data, Rename("data")); } //____________________________________ void DoHypothesisTest(RooWorkspace* wks){ // Use a RooStats ProfileLikleihoodCalculator to do the hypothesis test. ModelConfig model; model.SetWorkspace(*wks); model.SetPdf("model"); //plc.SetData("data"); ProfileLikelihoodCalculator plc; plc.SetData( *(wks->data("data") )); // here we explicitly set the value of the parameters for the null. // We want no signal contribution, eg. mu = 0 RooRealVar* mu = wks->var("mu"); // RooArgSet* nullParams = new RooArgSet("nullParams"); // nullParams->addClone(*mu); RooArgSet poi(*mu); RooArgSet * nullParams = (RooArgSet*) poi.snapshot(); nullParams->setRealValue("mu",0); //plc.SetNullParameters(*nullParams); plc.SetModel(model); // NOTE: using snapshot will import nullparams // in the WS and merge with existing "mu" // model.SetSnapshot(*nullParams); //use instead setNuisanceParameters plc.SetNullParameters( *nullParams); // We get a HypoTestResult out of the calculator, and we can query it. HypoTestResult* htr = plc.GetHypoTest(); cout << "-------------------------------------------------" << endl; cout << "The p-value for the null is " << htr->NullPValue() << endl; cout << "Corresponding to a signifcance of " << htr->Significance() << endl; cout << "-------------------------------------------------\n\n" << endl; } //____________________________________ void MakePlots(RooWorkspace* wks) { // Make plots of the data and the best fit model in two cases: // first the signal+background case // second the background-only case. // get some things out of workspace RooAbsPdf* model = wks->pdf("model"); RooAbsPdf* sigModel = wks->pdf("sigModel"); RooAbsPdf* zjjModel = wks->pdf("zjjModel"); RooAbsPdf* qcdModel = wks->pdf("qcdModel"); RooRealVar* mu = wks->var("mu"); RooRealVar* invMass = wks->var("invMass"); RooAbsData* data = wks->data("data"); ////////////////////////////////////////////////////////// // Make plots for the Alternate hypothesis, eg. let mu float mu->setConstant(kFALSE); model->fitTo(*data,Save(kTRUE),Minos(kFALSE), Hesse(kFALSE),PrintLevel(-1)); //plot sig candidates, full model, and individual componenets new TCanvas(); RooPlot* frame = invMass->frame() ; data->plotOn(frame ) ; model->plotOn(frame) ; model->plotOn(frame,Components(*sigModel),LineStyle(kDashed), LineColor(kRed)) ; model->plotOn(frame,Components(*zjjModel),LineStyle(kDashed),LineColor(kBlack)) ; model->plotOn(frame,Components(*qcdModel),LineStyle(kDashed),LineColor(kGreen)) ; frame->SetTitle("An example fit to the signal + background model"); frame->Draw() ; // cdata->SaveAs("alternateFit.gif"); ////////////////////////////////////////////////////////// // Do Fit to the Null hypothesis. Eg. fix mu=0 mu->setVal(0); // set signal fraction to 0 mu->setConstant(kTRUE); // set constant model->fitTo(*data, Save(kTRUE), Minos(kFALSE), Hesse(kFALSE),PrintLevel(-1)); // plot signal candidates with background model and components new TCanvas(); RooPlot* xframe2 = invMass->frame() ; data->plotOn(xframe2, DataError(RooAbsData::SumW2)) ; model->plotOn(xframe2) ; model->plotOn(xframe2, Components(*zjjModel),LineStyle(kDashed),LineColor(kBlack)) ; model->plotOn(xframe2, Components(*qcdModel),LineStyle(kDashed),LineColor(kGreen)) ; xframe2->SetTitle("An example fit to the background-only model"); xframe2->Draw() ; // cbkgonly->SaveAs("nullFit.gif"); } rs102_hypotestwithshapes.C:1 rs102_hypotestwithshapes.C:2 rs102_hypotestwithshapes.C:3 rs102_hypotestwithshapes.C:4 rs102_hypotestwithshapes.C:5 rs102_hypotestwithshapes.C:6 rs102_hypotestwithshapes.C:7 rs102_hypotestwithshapes.C:8 rs102_hypotestwithshapes.C:9 rs102_hypotestwithshapes.C:10 rs102_hypotestwithshapes.C:11 rs102_hypotestwithshapes.C:12 rs102_hypotestwithshapes.C:13 rs102_hypotestwithshapes.C:14 rs102_hypotestwithshapes.C:15 rs102_hypotestwithshapes.C:16 rs102_hypotestwithshapes.C:17 rs102_hypotestwithshapes.C:18 rs102_hypotestwithshapes.C:19 rs102_hypotestwithshapes.C:20 rs102_hypotestwithshapes.C:21 rs102_hypotestwithshapes.C:22 rs102_hypotestwithshapes.C:23 rs102_hypotestwithshapes.C:24 rs102_hypotestwithshapes.C:25 rs102_hypotestwithshapes.C:26 rs102_hypotestwithshapes.C:27 rs102_hypotestwithshapes.C:28 rs102_hypotestwithshapes.C:29 rs102_hypotestwithshapes.C:30 rs102_hypotestwithshapes.C:31 rs102_hypotestwithshapes.C:32 rs102_hypotestwithshapes.C:33 rs102_hypotestwithshapes.C:34 rs102_hypotestwithshapes.C:35 rs102_hypotestwithshapes.C:36 rs102_hypotestwithshapes.C:37 rs102_hypotestwithshapes.C:38 rs102_hypotestwithshapes.C:39 rs102_hypotestwithshapes.C:40 rs102_hypotestwithshapes.C:41 rs102_hypotestwithshapes.C:42 rs102_hypotestwithshapes.C:43 rs102_hypotestwithshapes.C:44 rs102_hypotestwithshapes.C:45 rs102_hypotestwithshapes.C:46 rs102_hypotestwithshapes.C:47 rs102_hypotestwithshapes.C:48 rs102_hypotestwithshapes.C:49 rs102_hypotestwithshapes.C:50 rs102_hypotestwithshapes.C:51 rs102_hypotestwithshapes.C:52 rs102_hypotestwithshapes.C:53 rs102_hypotestwithshapes.C:54 rs102_hypotestwithshapes.C:55 rs102_hypotestwithshapes.C:56 rs102_hypotestwithshapes.C:57 rs102_hypotestwithshapes.C:58 rs102_hypotestwithshapes.C:59 rs102_hypotestwithshapes.C:60 rs102_hypotestwithshapes.C:61 rs102_hypotestwithshapes.C:62 rs102_hypotestwithshapes.C:63 rs102_hypotestwithshapes.C:64 rs102_hypotestwithshapes.C:65 rs102_hypotestwithshapes.C:66 rs102_hypotestwithshapes.C:67 rs102_hypotestwithshapes.C:68 rs102_hypotestwithshapes.C:69 rs102_hypotestwithshapes.C:70 rs102_hypotestwithshapes.C:71 rs102_hypotestwithshapes.C:72 rs102_hypotestwithshapes.C:73 rs102_hypotestwithshapes.C:74 rs102_hypotestwithshapes.C:75 rs102_hypotestwithshapes.C:76 rs102_hypotestwithshapes.C:77 rs102_hypotestwithshapes.C:78 rs102_hypotestwithshapes.C:79 rs102_hypotestwithshapes.C:80 rs102_hypotestwithshapes.C:81 rs102_hypotestwithshapes.C:82 rs102_hypotestwithshapes.C:83 rs102_hypotestwithshapes.C:84 rs102_hypotestwithshapes.C:85 rs102_hypotestwithshapes.C:86 rs102_hypotestwithshapes.C:87 rs102_hypotestwithshapes.C:88 rs102_hypotestwithshapes.C:89 rs102_hypotestwithshapes.C:90 rs102_hypotestwithshapes.C:91 rs102_hypotestwithshapes.C:92 rs102_hypotestwithshapes.C:93 rs102_hypotestwithshapes.C:94 rs102_hypotestwithshapes.C:95 rs102_hypotestwithshapes.C:96 rs102_hypotestwithshapes.C:97 rs102_hypotestwithshapes.C:98 rs102_hypotestwithshapes.C:99 rs102_hypotestwithshapes.C:100 rs102_hypotestwithshapes.C:101 rs102_hypotestwithshapes.C:102 rs102_hypotestwithshapes.C:103 rs102_hypotestwithshapes.C:104 rs102_hypotestwithshapes.C:105 rs102_hypotestwithshapes.C:106 rs102_hypotestwithshapes.C:107 rs102_hypotestwithshapes.C:108 rs102_hypotestwithshapes.C:109 rs102_hypotestwithshapes.C:110 rs102_hypotestwithshapes.C:111 rs102_hypotestwithshapes.C:112 rs102_hypotestwithshapes.C:113 rs102_hypotestwithshapes.C:114 rs102_hypotestwithshapes.C:115 rs102_hypotestwithshapes.C:116 rs102_hypotestwithshapes.C:117 rs102_hypotestwithshapes.C:118 rs102_hypotestwithshapes.C:119 rs102_hypotestwithshapes.C:120 rs102_hypotestwithshapes.C:121 rs102_hypotestwithshapes.C:122 rs102_hypotestwithshapes.C:123 rs102_hypotestwithshapes.C:124 rs102_hypotestwithshapes.C:125 rs102_hypotestwithshapes.C:126 rs102_hypotestwithshapes.C:127 rs102_hypotestwithshapes.C:128 rs102_hypotestwithshapes.C:129 rs102_hypotestwithshapes.C:130 rs102_hypotestwithshapes.C:131 rs102_hypotestwithshapes.C:132 rs102_hypotestwithshapes.C:133 rs102_hypotestwithshapes.C:134 rs102_hypotestwithshapes.C:135 rs102_hypotestwithshapes.C:136 rs102_hypotestwithshapes.C:137 rs102_hypotestwithshapes.C:138 rs102_hypotestwithshapes.C:139 rs102_hypotestwithshapes.C:140 rs102_hypotestwithshapes.C:141 rs102_hypotestwithshapes.C:142 rs102_hypotestwithshapes.C:143 rs102_hypotestwithshapes.C:144 rs102_hypotestwithshapes.C:145 rs102_hypotestwithshapes.C:146 rs102_hypotestwithshapes.C:147 rs102_hypotestwithshapes.C:148 rs102_hypotestwithshapes.C:149 rs102_hypotestwithshapes.C:150 rs102_hypotestwithshapes.C:151 rs102_hypotestwithshapes.C:152 rs102_hypotestwithshapes.C:153 rs102_hypotestwithshapes.C:154 rs102_hypotestwithshapes.C:155 rs102_hypotestwithshapes.C:156 rs102_hypotestwithshapes.C:157 rs102_hypotestwithshapes.C:158 rs102_hypotestwithshapes.C:159 rs102_hypotestwithshapes.C:160 rs102_hypotestwithshapes.C:161 rs102_hypotestwithshapes.C:162 rs102_hypotestwithshapes.C:163 rs102_hypotestwithshapes.C:164 rs102_hypotestwithshapes.C:165 rs102_hypotestwithshapes.C:166 rs102_hypotestwithshapes.C:167 rs102_hypotestwithshapes.C:168 rs102_hypotestwithshapes.C:169 rs102_hypotestwithshapes.C:170 rs102_hypotestwithshapes.C:171 rs102_hypotestwithshapes.C:172 rs102_hypotestwithshapes.C:173 rs102_hypotestwithshapes.C:174 rs102_hypotestwithshapes.C:175 rs102_hypotestwithshapes.C:176 rs102_hypotestwithshapes.C:177 rs102_hypotestwithshapes.C:178 rs102_hypotestwithshapes.C:179 rs102_hypotestwithshapes.C:180 rs102_hypotestwithshapes.C:181 rs102_hypotestwithshapes.C:182 rs102_hypotestwithshapes.C:183 rs102_hypotestwithshapes.C:184 rs102_hypotestwithshapes.C:185 rs102_hypotestwithshapes.C:186 rs102_hypotestwithshapes.C:187 rs102_hypotestwithshapes.C:188 rs102_hypotestwithshapes.C:189 rs102_hypotestwithshapes.C:190 rs102_hypotestwithshapes.C:191 rs102_hypotestwithshapes.C:192 rs102_hypotestwithshapes.C:193 rs102_hypotestwithshapes.C:194 rs102_hypotestwithshapes.C:195 rs102_hypotestwithshapes.C:196 rs102_hypotestwithshapes.C:197 rs102_hypotestwithshapes.C:198 rs102_hypotestwithshapes.C:199 rs102_hypotestwithshapes.C:200 rs102_hypotestwithshapes.C:201 rs102_hypotestwithshapes.C:202 rs102_hypotestwithshapes.C:203 rs102_hypotestwithshapes.C:204 rs102_hypotestwithshapes.C:205 rs102_hypotestwithshapes.C:206 rs102_hypotestwithshapes.C:207 rs102_hypotestwithshapes.C:208 rs102_hypotestwithshapes.C:209 rs102_hypotestwithshapes.C:210 rs102_hypotestwithshapes.C:211 rs102_hypotestwithshapes.C:212 rs102_hypotestwithshapes.C:213 rs102_hypotestwithshapes.C:214 rs102_hypotestwithshapes.C:215 rs102_hypotestwithshapes.C:216 rs102_hypotestwithshapes.C:217 rs102_hypotestwithshapes.C:218 rs102_hypotestwithshapes.C:219 rs102_hypotestwithshapes.C:220 rs102_hypotestwithshapes.C:221 rs102_hypotestwithshapes.C:222 rs102_hypotestwithshapes.C:223 rs102_hypotestwithshapes.C:224 rs102_hypotestwithshapes.C:225 rs102_hypotestwithshapes.C:226 rs102_hypotestwithshapes.C:227 rs102_hypotestwithshapes.C:228 rs102_hypotestwithshapes.C:229 rs102_hypotestwithshapes.C:230 rs102_hypotestwithshapes.C:231 rs102_hypotestwithshapes.C:232 rs102_hypotestwithshapes.C:233 rs102_hypotestwithshapes.C:234 rs102_hypotestwithshapes.C:235 rs102_hypotestwithshapes.C:236 rs102_hypotestwithshapes.C:237 rs102_hypotestwithshapes.C:238 rs102_hypotestwithshapes.C:239 rs102_hypotestwithshapes.C:240 rs102_hypotestwithshapes.C:241 rs102_hypotestwithshapes.C:242 rs102_hypotestwithshapes.C:243 rs102_hypotestwithshapes.C:244 rs102_hypotestwithshapes.C:245 rs102_hypotestwithshapes.C:246 rs102_hypotestwithshapes.C:247 rs102_hypotestwithshapes.C:248 rs102_hypotestwithshapes.C:249 rs102_hypotestwithshapes.C:250 rs102_hypotestwithshapes.C:251 rs102_hypotestwithshapes.C:252 rs102_hypotestwithshapes.C:253 rs102_hypotestwithshapes.C:254 rs102_hypotestwithshapes.C:255 rs102_hypotestwithshapes.C:256 rs102_hypotestwithshapes.C:257 rs102_hypotestwithshapes.C:258 rs102_hypotestwithshapes.C:259 rs102_hypotestwithshapes.C:260 rs102_hypotestwithshapes.C:261 rs102_hypotestwithshapes.C:262 rs102_hypotestwithshapes.C:263 rs102_hypotestwithshapes.C:264 rs102_hypotestwithshapes.C:265 rs102_hypotestwithshapes.C:266 rs102_hypotestwithshapes.C:267 rs102_hypotestwithshapes.C:268 rs102_hypotestwithshapes.C:269 rs102_hypotestwithshapes.C:270 rs102_hypotestwithshapes.C:271 rs102_hypotestwithshapes.C:272 rs102_hypotestwithshapes.C:273 rs102_hypotestwithshapes.C:274 rs102_hypotestwithshapes.C:275 rs102_hypotestwithshapes.C:276 rs102_hypotestwithshapes.C:277 rs102_hypotestwithshapes.C:278 rs102_hypotestwithshapes.C:279 |
|