This tutorial produces an N-dimensional multivariate Gaussian with a non-trivial covariance matrix. By default N=4 (called "dim").
A subset of these are considered parameters of interest. This problem is tractable analytically.
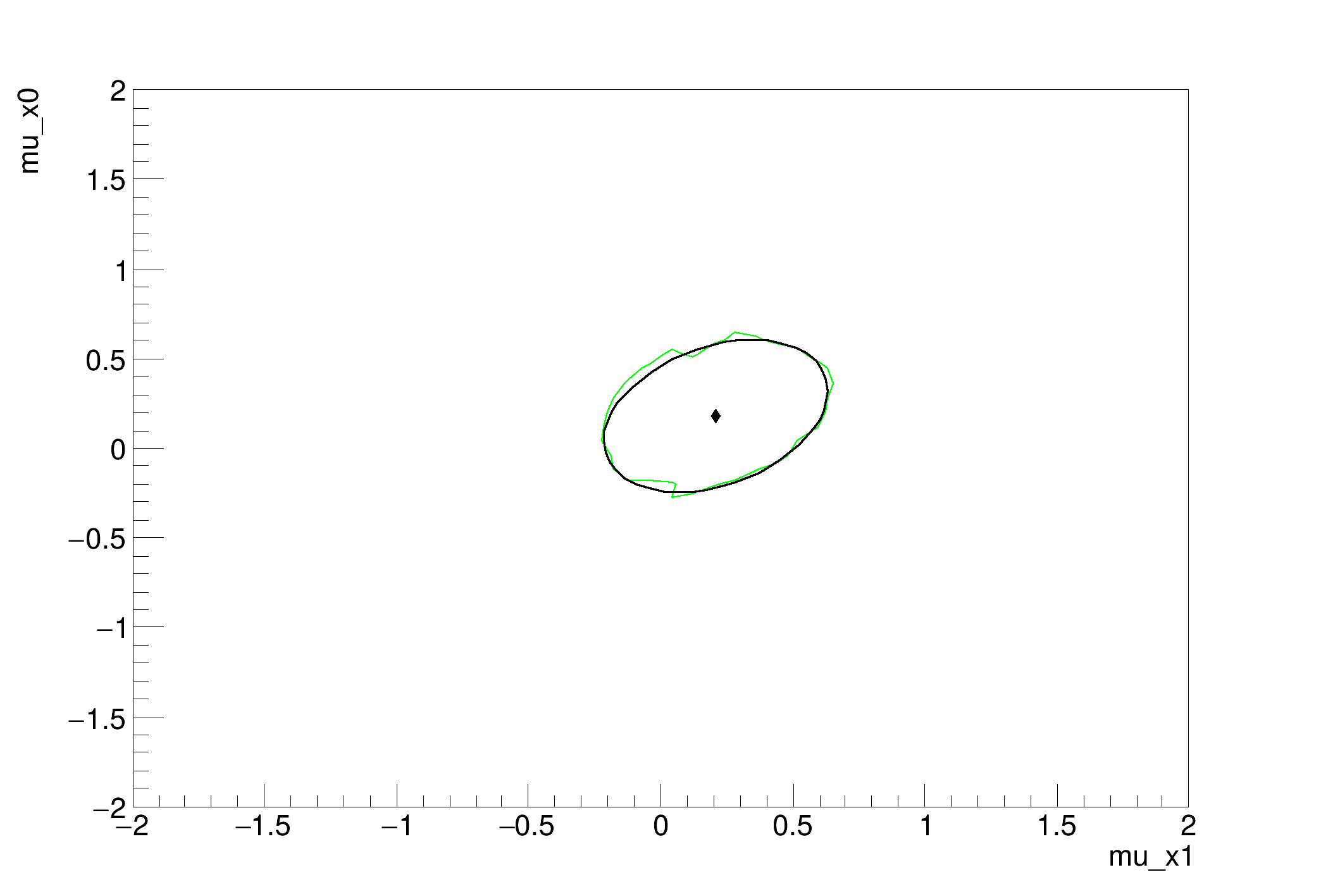
We use this mainly as a test of Markov Chain Monte Carlo and we compare the result to the profile likelihood ratio.
We use the proposal helper to create a customized proposal function for this problem.
For N=4 and 2 parameters of interest it takes about 10-20 seconds and the acceptance rate is 37%
[#1] INFO:Fitting -- RooAbsPdf::fitTo(mvg_over_mvg_Int[x0,x1,x2,x3]) fixing normalization set for coefficient determination to observables in data
[#1] INFO:Fitting -- using generic CPU library compiled with no vectorizations
[#1] INFO:Fitting -- Creation of NLL object took 7.1452 ms
[#1] INFO:Fitting -- RooAddition::defaultErrorLevel(nll_mvg_over_mvg_Int[x0,x1,x2,x3]_mvgData) Summation contains a RooNLLVar, using its error level
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: activating const optimization
[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
Minuit2Minimizer: Minimize with max-calls 2000 convergence for edm < 1 strategy 1
Minuit2Minimizer : Valid minimum - status = 0
FVAL = 706.560063684865781
Edm = 9.79361278577427602e-06
Nfcn = 68
mu_x0 = 0.180728 +/- 0.17298 (limited)
mu_x1 = 0.207351 +/- 0.172978 (limited)
mu_x2 = -0.0159412 +/- 0.172984 (limited)
mu_x3 = 0.12343 +/- 0.172982 (limited)
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: deactivating const optimization
[#1] INFO:Fitting -- RooAbsPdf::fitTo(mvg_over_mvg_Int[x0,x1,x2,x3]) fixing normalization set for coefficient determination to observables in data
[#1] INFO:Fitting -- Creation of NLL object took 164.2 μs
Metropolis-Hastings progress: ....................................................................................................
[#1] INFO:Eval -- Proposal acceptance rate: 37.1%
[#1] INFO:Eval -- Number of steps in chain: 3710
[#1] INFO:InputArguments -- The deprecated RooFit::CloneData(1) option passed to createNLL() is ignored.
[#1] INFO:Fitting -- RooAbsPdf::fitTo(mvg_over_mvg_Int[x0,x1,x2,x3]) fixing normalization set for coefficient determination to observables in data
[#1] INFO:Fitting -- Creation of NLL object took 141.011 μs
[#0] PROGRESS:Minimization -- ProfileLikelihoodCalcultor::DoGLobalFit - find MLE
[#1] INFO:Fitting -- RooAddition::defaultErrorLevel(nll_mvg_over_mvg_Int[x0,x1,x2,x3]_mvgData) Summation contains a RooNLLVar, using its error level
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: activating const optimization
[#0] PROGRESS:Minimization -- ProfileLikelihoodCalcultor::DoMinimizeNLL - using Minuit2 / Migrad with strategy 1
[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
[#1] INFO:Minimization --
RooFitResult: minimized FCN value: 706.56, estimated distance to minimum: 2.41067e-06
covariance matrix quality: Full, accurate covariance matrix
Status : MINIMIZE=0
Floating Parameter FinalValue +/- Error
-------------------- --------------------------
mu_x0 1.8134e-01 +/- 1.73e-01
mu_x1 2.0772e-01 +/- 1.73e-01
mu_x2 -1.5923e-02 +/- 1.73e-01
mu_x3 1.2360e-01 +/- 1.73e-01
[#1] INFO:Minimization -- RooProfileLL::evaluate(RooEvaluatorWrapper_Profile[mu_x0,mu_x1]) Creating instance of MINUIT
[#1] INFO:Fitting -- RooAddition::defaultErrorLevel(nll_mvg_over_mvg_Int[x0,x1,x2,x3]_mvgData) Summation contains a RooNLLVar, using its error level
[#1] INFO:Minimization -- RooProfileLL::evaluate(RooEvaluatorWrapper_Profile[mu_x0,mu_x1]) determining minimum likelihood for current configurations w.r.t all observable
[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
[#1] INFO:Minimization -- RooProfileLL::evaluate(RooEvaluatorWrapper_Profile[mu_x0,mu_x1]) minimum found at (mu_x0=0.181185, mu_x1=0.207918)
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
[#1] INFO:Minimization -- LikelihoodInterval - Finding the contour of mu_x0 ( 0 ) and mu_x1 ( 1 )
MCMC interval on p0: [-0.28, 0.6000000000000001]
MCMC interval on p1: [-0.19999999999999996, 0.6000000000000001]
Real time 0:00:01, CP time 1.040