
 Standard demo of the Bayesian MCMC calculator
Standard demo of the Bayesian MCMC calculator
This is a standard demo that can be used with any ROOT file prepared in the standard way. You specify:
- name for input ROOT file
- name of workspace inside ROOT file that holds model and data
- name of ModelConfig that specifies details for calculator tools
- name of dataset
With default parameters the macro will attempt to run the standard hist2workspace example and read the ROOT file that it produces.
The actual heart of the demo is only about 10 lines long.
The MCMCCalculator is a Bayesian tool that uses the Metropolis-Hastings algorithm to efficiently integrate in many dimensions. It is not as accurate as the BayesianCalculator for simple problems, but it scales to much more complicated cases.
Processing /mnt/build/workspace/root-makedoc-v612/rootspi/rdoc/src/v6-12-00-patches/tutorials/roostats/StandardBayesianMCMCDemo.C...
�[1mRooFit v3.60 -- Developed by Wouter Verkerke and David Kirkby�[0m
Copyright (C) 2000-2013 NIKHEF, University of California & Stanford University
All rights reserved, please read http://roofit.sourceforge.net/license.txt
[#1] INFO:Minization -- p.d.f. provides expected number of events, including extended term in likelihood.
[#1] INFO:Minization -- createNLL: caching constraint set under name CONSTR_OF_PDF_simPdf_FOR_OBS_channelCat:obs_x_channel1 with 6 entries
[#1] INFO:Minization -- Including the following contraint terms in minimization: (lumiConstraint,alpha_syst1Constraint,alpha_syst2Constraint,alpha_syst3Constraint,gamma_stat_channel1_bin_0_constraint,gamma_stat_channel1_bin_1_constraint)
[#1] INFO:Minization -- The following global observables have been defined: (nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
Metropolis-Hastings progress: ....................................................................................................
[#1] INFO:Eval -- Proposal acceptance rate: 27.377%
[#1] INFO:Eval -- Number of steps in chain: 27377
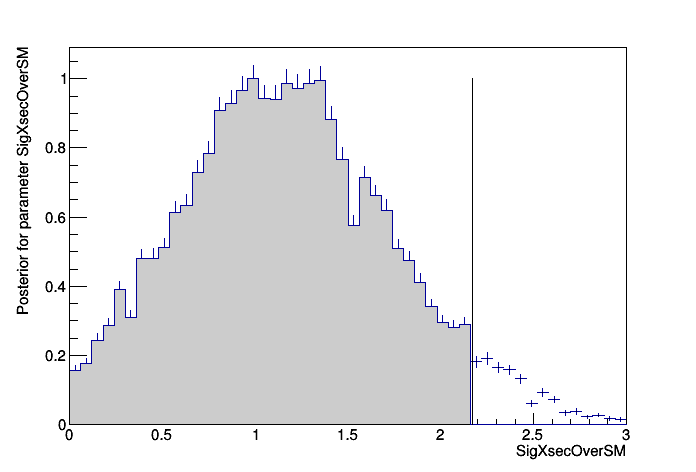
>>>> RESULT : 95% interval on SigXsecOverSM is : [0, 2.17276]
struct BayesianMCMCOptions {
double confLevel = 0.95;
int intervalType = 2;
double maxPOI = -999;
double minPOI = -999;
int numIters = 100000;
int numBurnInSteps = 100;
};
BayesianMCMCOptions optMCMC;
void StandardBayesianMCMCDemo(const char* infile = "",
const char* workspaceName = "combined",
const char* modelConfigName = "ModelConfig",
const char* dataName = "obsData"){
const char* filename = "";
if (!strcmp(infile,"")) {
filename = "results/example_combined_GaussExample_model.root";
if (!fileExist) {
#ifdef _WIN32
cout << "HistFactory file cannot be generated on Windows - exit" << endl;
return;
#endif
cout <<"will run standard hist2workspace example"<<endl;
gROOT->ProcessLine(
".! prepareHistFactory .");
gROOT->ProcessLine(
".! hist2workspace config/example.xml");
cout <<"\n\n---------------------"<<endl;
cout <<"Done creating example input"<<endl;
cout <<"---------------------\n\n"<<endl;
}
}
else
filename = infile;
if(!file ){
cout <<"StandardRooStatsDemoMacro: Input file " << filename << " is not found" << endl;
return;
}
if(!w){
cout <<"workspace not found" << endl;
return;
}
if(!data || !mc){
cout << "data or ModelConfig was not found" <<endl;
return;
}
mcmc.SetConfidenceLevel(optMCMC.confLevel);
mcmc.SetProposalFunction(sp);
mcmc.SetNumIters(optMCMC.numIters);
mcmc.SetNumBurnInSteps(optMCMC.numBurnInSteps);
if (optMCMC.intervalType == 0) mcmc.SetIntervalType(MCMCInterval::kShortest);
if (optMCMC.intervalType == 1) mcmc.SetLeftSideTailFraction(0.5);
if (optMCMC.intervalType == 2) mcmc.SetLeftSideTailFraction(0.);
if (optMCMC.minPOI != -999)
firstPOI->
setMin(optMCMC.minPOI);
if (optMCMC.maxPOI != -999)
firstPOI->
setMax(optMCMC.maxPOI);
auto c1 =
new TCanvas(
"IntervalPlot");
plot.Draw();
}
int iPad=1;
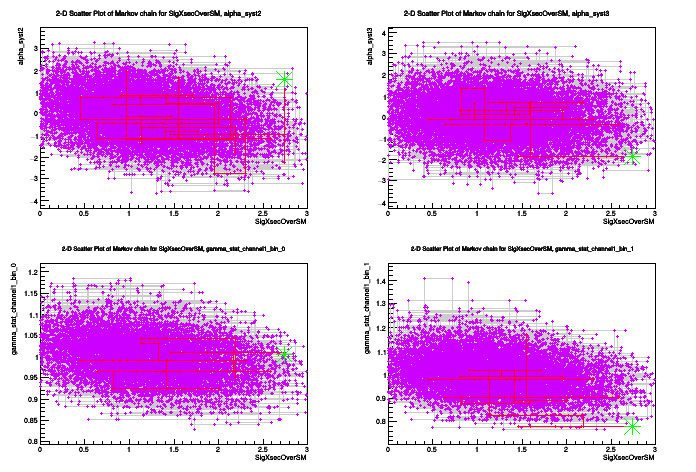
plot.DrawChainScatter(*firstPOI,*nuis);
}
cout <<
"\n>>>> RESULT : " << optMCMC.confLevel*100 <<
"% interval on " <<firstPOI->
GetName()<<
" is : ["<<
}
- Author
- Kyle Cranmer
Definition in file StandardBayesianMCMCDemo.C.



 Standard demo of the Bayesian MCMC calculator
Standard demo of the Bayesian MCMC calculator 
