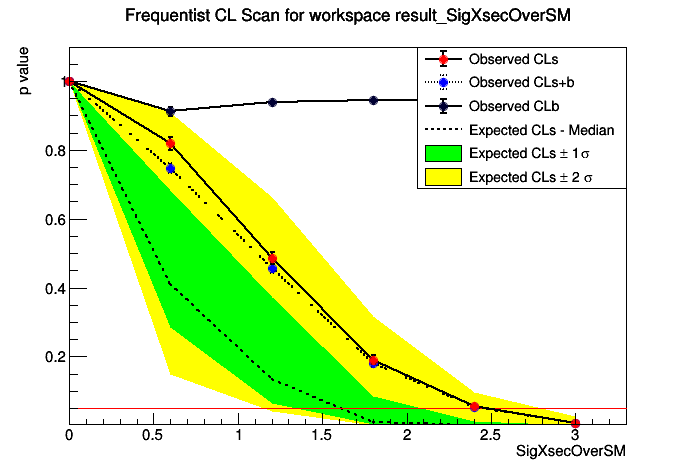
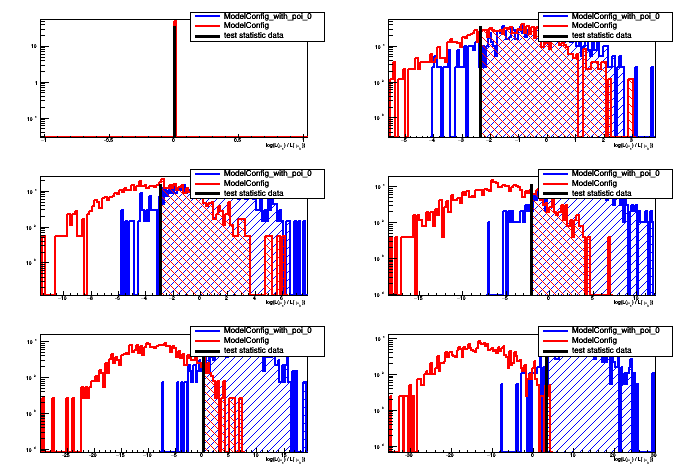
This macro will perform a scan of the p-values for computing the interval or limit
Processing /mnt/build/workspace/root-makedoc-v612/rootspi/rdoc/src/v6-12-00-patches/tutorials/roostats/StandardHypoTestInvDemo.C...
�[1mRooFit v3.60 -- Developed by Wouter Verkerke and David Kirkby�[0m
Copyright (C) 2000-2013 NIKHEF, University of California & Stanford University
All rights reserved, please read http://roofit.sourceforge.net/license.txt
0x3535760 results/example_combined_GaussExample_model.root
Running HypoTestInverter on the workspace combined
RooWorkspace(combined) combined contents
variables
---------
(Lumi,SigXsecOverSM,alpha_syst1,alpha_syst2,alpha_syst3,binWidth_obs_x_channel1_0,binWidth_obs_x_channel1_1,binWidth_obs_x_channel1_2,channelCat,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1,nom_alpha_syst1,nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1,nominalLumi,obs_x_channel1,weightVar)
p.d.f.s
-------
RooGaussian::alpha_syst1Constraint[ x=alpha_syst1 mean=nom_alpha_syst1 sigma=1 ] = 1
RooGaussian::alpha_syst2Constraint[ x=alpha_syst2 mean=nom_alpha_syst2 sigma=1 ] = 1
RooGaussian::alpha_syst3Constraint[ x=alpha_syst3 mean=nom_alpha_syst3 sigma=1 ] = 1
RooRealSumPdf::channel1_model[ binWidth_obs_x_channel1_0 * L_x_signal_channel1_overallSyst_x_Exp + binWidth_obs_x_channel1_1 * L_x_background1_channel1_overallSyst_x_StatUncert + binWidth_obs_x_channel1_2 * L_x_background2_channel1_overallSyst_x_StatUncert ] = 220
RooPoisson::gamma_stat_channel1_bin_0_constraint[ x=nom_gamma_stat_channel1_bin_0 mean=gamma_stat_channel1_bin_0_poisMean ] = 0.019943
RooPoisson::gamma_stat_channel1_bin_1_constraint[ x=nom_gamma_stat_channel1_bin_1 mean=gamma_stat_channel1_bin_1_poisMean ] = 0.039861
RooGaussian::lumiConstraint[ x=Lumi mean=nominalLumi sigma=0.1 ] = 1
RooProdPdf::model_channel1[ lumiConstraint * alpha_syst1Constraint * alpha_syst2Constraint * alpha_syst3Constraint * gamma_stat_channel1_bin_0_constraint * gamma_stat_channel1_bin_1_constraint * channel1_model(obs_x_channel1) ] = 0.174888
RooSimultaneous::simPdf[ indexCat=channelCat channel1=model_channel1 ] = 0.174888
functions
--------
RooProduct::L_x_background1_channel1_overallSyst_x_StatUncert[ Lumi * background1_channel1_overallSyst_x_StatUncert ] = 0
RooProduct::L_x_background2_channel1_overallSyst_x_StatUncert[ Lumi * background2_channel1_overallSyst_x_StatUncert ] = 100
RooProduct::L_x_signal_channel1_overallSyst_x_Exp[ Lumi * signal_channel1_overallSyst_x_Exp ] = 10
RooStats::HistFactory::FlexibleInterpVar::background1_channel1_epsilon[ paramList=(alpha_syst2) ] = 1
RooHistFunc::background1_channel1_nominal[ depList=(obs_x_channel1) depList=(obs_x_channel1) ] = 0
RooProduct::background1_channel1_overallSyst_x_Exp[ background1_channel1_nominal * background1_channel1_epsilon ] = 0
RooProduct::background1_channel1_overallSyst_x_StatUncert[ mc_stat_channel1 * background1_channel1_overallSyst_x_Exp ] = 0
RooStats::HistFactory::FlexibleInterpVar::background2_channel1_epsilon[ paramList=(alpha_syst3) ] = 1
RooHistFunc::background2_channel1_nominal[ depList=(obs_x_channel1) depList=(obs_x_channel1) ] = 100
RooProduct::background2_channel1_overallSyst_x_Exp[ background2_channel1_nominal * background2_channel1_epsilon ] = 100
RooProduct::background2_channel1_overallSyst_x_StatUncert[ mc_stat_channel1 * background2_channel1_overallSyst_x_Exp ] = 100
RooProduct::gamma_stat_channel1_bin_0_poisMean[ gamma_stat_channel1_bin_0 * gamma_stat_channel1_bin_0_tau ] = 400
RooProduct::gamma_stat_channel1_bin_1_poisMean[ gamma_stat_channel1_bin_1 * gamma_stat_channel1_bin_1_tau ] = 100
ParamHistFunc::mc_stat_channel1[ ] = 1
RooStats::HistFactory::FlexibleInterpVar::signal_channel1_epsilon[ paramList=(alpha_syst1) ] = 1
RooHistFunc::signal_channel1_nominal[ depList=(obs_x_channel1) depList=(obs_x_channel1) ] = 10
RooProduct::signal_channel1_overallNorm_x_sigma_epsilon[ SigXsecOverSM * signal_channel1_epsilon ] = 1
RooProduct::signal_channel1_overallSyst_x_Exp[ signal_channel1_nominal * signal_channel1_overallNorm_x_sigma_epsilon ] = 10
datasets
--------
RooDataSet::asimovData(obs_x_channel1,weightVar,channelCat)
RooDataSet::obsData(channelCat,obs_x_channel1)
embedded datasets (in pdfs and functions)
-----------------------------------------
RooDataHist::signal_channel1nominalDHist(obs_x_channel1)
RooDataHist::background1_channel1nominalDHist(obs_x_channel1)
RooDataHist::background2_channel1nominalDHist(obs_x_channel1)
parameter snapshots
-------------------
NominalParamValues = (nom_alpha_syst2=0[C],nom_alpha_syst3=0[C],nom_gamma_stat_channel1_bin_0=400[C],nom_gamma_stat_channel1_bin_1=100[C],weightVar=0,obs_x_channel1=1.75,Lumi=1[C],nominalLumi=1[C],alpha_syst1=0[C],nom_alpha_syst1=0[C],alpha_syst2=0,alpha_syst3=0,gamma_stat_channel1_bin_0=1,gamma_stat_channel1_bin_1=1,SigXsecOverSM=1,binWidth_obs_x_channel1_0=2[C],binWidth_obs_x_channel1_1=2[C],binWidth_obs_x_channel1_2=2[C])
named sets
----------
ModelConfig_GlobalObservables:(nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
ModelConfig_NuisParams:(alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
ModelConfig_Observables:(obs_x_channel1,weightVar,channelCat)
ModelConfig_POI:(SigXsecOverSM)
globalObservables:(nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
observables:(obs_x_channel1,weightVar,channelCat)
generic objects
---------------
RooStats::ModelConfig::ModelConfig
Using data set obsData
StandardHypoTestInvDemo : POI initial value: SigXsecOverSM = 1
[#1] INFO:InputArguments -- HypoTestInverter ---- Input models:
using as S+B (null) model : ModelConfig
using as B (alternate) model : ModelConfig_with_poi_0
Doing a fixed scan in interval : 0 , 5
[#1] INFO:Eval -- HypoTestInverter::GetInterval - run a fixed scan
[#0] WARNING:InputArguments -- HypoTestInverter::RunFixedScan - xMax > upper bound, use xmax = 3
[#1] INFO:ObjectHandling -- RooWorkspace::saveSnaphot(combined) replacing previous snapshot with name ModelConfig__snapshot
[#0] PROGRESS:Eval -- Running for SigXsecOverSM = 0
=== Using the following for ModelConfig ===
Observables: RooArgSet:: = (obs_x_channel1,weightVar,channelCat)
Parameters of Interest: RooArgSet:: = (SigXsecOverSM)
Nuisance Parameters: RooArgSet:: = (alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
Global Observables: RooArgSet:: = (nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
PDF: RooSimultaneous::simPdf[ indexCat=channelCat channel1=model_channel1 ] = 0.158989
Snapshot:
1) 0x43cda40 RooRealVar:: SigXsecOverSM = 0 L(0 - 3) "SigXsecOverSM"
=== Using the following for ModelConfig_with_poi_0 ===
Observables: RooArgSet:: = (obs_x_channel1,weightVar,channelCat)
Parameters of Interest: RooArgSet:: = (SigXsecOverSM)
Nuisance Parameters: RooArgSet:: = (alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
Global Observables: RooArgSet:: = (nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
PDF: RooSimultaneous::simPdf[ indexCat=channelCat channel1=model_channel1 ] = 0.158989
Snapshot:
1) 0x43cda40 RooRealVar:: SigXsecOverSM = 0 L(0 - 3) "SigXsecOverSM"
[#0] PROGRESS:Generation -- Test Statistic on data: 0
[#1] INFO:InputArguments -- Profiling conditional MLEs for Null.
[#1] INFO:InputArguments -- Using a ToyMCSampler. Now configuring for Null.
[#0] PROGRESS:Generation -- generated toys: 500 / 1000
[#1] INFO:InputArguments -- Profiling conditional MLEs for Alt.
[#1] INFO:InputArguments -- Using a ToyMCSampler. Now configuring for Alt.
[#0] PROGRESS:Eval -- P values for SigXsecOverSM = 0
CLs = 1 +/- 0
CLb = 1 +/- 0
CLsplusb = 1 +/- 0
[#1] INFO:ObjectHandling -- RooWorkspace::saveSnaphot(combined) replacing previous snapshot with name ModelConfig__snapshot
[#0] PROGRESS:Eval -- Running for SigXsecOverSM = 0.6
=== Using the following for ModelConfig ===
Observables: RooArgSet:: = (obs_x_channel1,weightVar,channelCat)
Parameters of Interest: RooArgSet:: = (SigXsecOverSM)
Nuisance Parameters: RooArgSet:: = (alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
Global Observables: RooArgSet:: = (nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
PDF: RooSimultaneous::simPdf[ indexCat=channelCat channel1=model_channel1 ] = 0.168529
Snapshot:
1) 0x49bf580 RooRealVar:: SigXsecOverSM = 0.6 L(0 - 3) "SigXsecOverSM"
=== Using the following for ModelConfig_with_poi_0 ===
Observables: RooArgSet:: = (obs_x_channel1,weightVar,channelCat)
Parameters of Interest: RooArgSet:: = (SigXsecOverSM)
Nuisance Parameters: RooArgSet:: = (alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
Global Observables: RooArgSet:: = (nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
PDF: RooSimultaneous::simPdf[ indexCat=channelCat channel1=model_channel1 ] = 0.168529
Snapshot:
1) 0x49bf580 RooRealVar:: SigXsecOverSM = 0 L(0 - 3) "SigXsecOverSM"
[#0] PROGRESS:Generation -- Test Statistic on data: -2.35222
[#1] INFO:InputArguments -- Profiling conditional MLEs for Null.
[#1] INFO:InputArguments -- Using a ToyMCSampler. Now configuring for Null.
[#0] PROGRESS:Generation -- generated toys: 500 / 1000
[#1] INFO:InputArguments -- Profiling conditional MLEs for Alt.
[#1] INFO:InputArguments -- Using a ToyMCSampler. Now configuring for Alt.
[#0] PROGRESS:Eval -- P values for SigXsecOverSM = 0.6
CLs = 0.819079 +/- 0.0188863
CLb = 0.912 +/- 0.0126693
CLsplusb = 0.747 +/- 0.0137474
[#1] INFO:ObjectHandling -- RooWorkspace::saveSnaphot(combined) replacing previous snapshot with name ModelConfig__snapshot
[#0] PROGRESS:Eval -- Running for SigXsecOverSM = 1.2
=== Using the following for ModelConfig ===
Observables: RooArgSet:: = (obs_x_channel1,weightVar,channelCat)
Parameters of Interest: RooArgSet:: = (SigXsecOverSM)
Nuisance Parameters: RooArgSet:: = (alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
Global Observables: RooArgSet:: = (nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
PDF: RooSimultaneous::simPdf[ indexCat=channelCat channel1=model_channel1 ] = 0.178068
Snapshot:
1) 0x49cb540 RooRealVar:: SigXsecOverSM = 1.2 L(0 - 3) "SigXsecOverSM"
=== Using the following for ModelConfig_with_poi_0 ===
Observables: RooArgSet:: = (obs_x_channel1,weightVar,channelCat)
Parameters of Interest: RooArgSet:: = (SigXsecOverSM)
Nuisance Parameters: RooArgSet:: = (alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
Global Observables: RooArgSet:: = (nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
PDF: RooSimultaneous::simPdf[ indexCat=channelCat channel1=model_channel1 ] = 0.178068
Snapshot:
1) 0x49cb540 RooRealVar:: SigXsecOverSM = 0 L(0 - 3) "SigXsecOverSM"
[#0] PROGRESS:Generation -- Test Statistic on data: -2.9364
[#1] INFO:InputArguments -- Profiling conditional MLEs for Null.
[#1] INFO:InputArguments -- Using a ToyMCSampler. Now configuring for Null.
[#0] PROGRESS:Generation -- generated toys: 500 / 1000
[#1] INFO:InputArguments -- Profiling conditional MLEs for Alt.
[#1] INFO:InputArguments -- Using a ToyMCSampler. Now configuring for Alt.
[#0] PROGRESS:Eval -- P values for SigXsecOverSM = 1.2
CLs = 0.48617 +/- 0.0176356
CLb = 0.94 +/- 0.0106207
CLsplusb = 0.457 +/- 0.0157528
[#1] INFO:ObjectHandling -- RooWorkspace::saveSnaphot(combined) replacing previous snapshot with name ModelConfig__snapshot
[#0] PROGRESS:Eval -- Running for SigXsecOverSM = 1.8
=== Using the following for ModelConfig ===
Observables: RooArgSet:: = (obs_x_channel1,weightVar,channelCat)
Parameters of Interest: RooArgSet:: = (SigXsecOverSM)
Nuisance Parameters: RooArgSet:: = (alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
Global Observables: RooArgSet:: = (nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
PDF: RooSimultaneous::simPdf[ indexCat=channelCat channel1=model_channel1 ] = 0.187607
Snapshot:
1) 0x48b33d0 RooRealVar:: SigXsecOverSM = 1.8 L(0 - 3) "SigXsecOverSM"
=== Using the following for ModelConfig_with_poi_0 ===
Observables: RooArgSet:: = (obs_x_channel1,weightVar,channelCat)
Parameters of Interest: RooArgSet:: = (SigXsecOverSM)
Nuisance Parameters: RooArgSet:: = (alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
Global Observables: RooArgSet:: = (nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
PDF: RooSimultaneous::simPdf[ indexCat=channelCat channel1=model_channel1 ] = 0.187607
Snapshot:
1) 0x48b33d0 RooRealVar:: SigXsecOverSM = 0 L(0 - 3) "SigXsecOverSM"
[#0] PROGRESS:Generation -- Test Statistic on data: -2.05075
[#1] INFO:InputArguments -- Profiling conditional MLEs for Null.
[#1] INFO:InputArguments -- Using a ToyMCSampler. Now configuring for Null.
[#0] PROGRESS:Generation -- generated toys: 500 / 1000
[#1] INFO:InputArguments -- Profiling conditional MLEs for Alt.
[#1] INFO:InputArguments -- Using a ToyMCSampler. Now configuring for Alt.
[#0] PROGRESS:Eval -- P values for SigXsecOverSM = 1.8
CLs = 0.190678 +/- 0.0130363
CLb = 0.944 +/- 0.0102824
CLsplusb = 0.18 +/- 0.0121491
[#1] INFO:ObjectHandling -- RooWorkspace::saveSnaphot(combined) replacing previous snapshot with name ModelConfig__snapshot
[#0] PROGRESS:Eval -- Running for SigXsecOverSM = 2.4
=== Using the following for ModelConfig ===
Observables: RooArgSet:: = (obs_x_channel1,weightVar,channelCat)
Parameters of Interest: RooArgSet:: = (SigXsecOverSM)
Nuisance Parameters: RooArgSet:: = (alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
Global Observables: RooArgSet:: = (nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
PDF: RooSimultaneous::simPdf[ indexCat=channelCat channel1=model_channel1 ] = 0.197147
Snapshot:
1) 0x49bf580 RooRealVar:: SigXsecOverSM = 2.4 L(0 - 3) "SigXsecOverSM"
=== Using the following for ModelConfig_with_poi_0 ===
Observables: RooArgSet:: = (obs_x_channel1,weightVar,channelCat)
Parameters of Interest: RooArgSet:: = (SigXsecOverSM)
Nuisance Parameters: RooArgSet:: = (alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
Global Observables: RooArgSet:: = (nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
PDF: RooSimultaneous::simPdf[ indexCat=channelCat channel1=model_channel1 ] = 0.197147
Snapshot:
1) 0x49bf580 RooRealVar:: SigXsecOverSM = 0 L(0 - 3) "SigXsecOverSM"
[#0] PROGRESS:Generation -- Test Statistic on data: 0.0783908
[#1] INFO:InputArguments -- Profiling conditional MLEs for Null.
[#1] INFO:InputArguments -- Using a ToyMCSampler. Now configuring for Null.
[#0] PROGRESS:Generation -- generated toys: 500 / 1000
[#1] INFO:InputArguments -- Profiling conditional MLEs for Alt.
[#1] INFO:InputArguments -- Using a ToyMCSampler. Now configuring for Alt.
[#0] PROGRESS:Eval -- P values for SigXsecOverSM = 2.4
CLs = 0.0550847 +/- 0.00746178
CLb = 0.944 +/- 0.0102824
CLsplusb = 0.052 +/- 0.00702111
[#1] INFO:ObjectHandling -- RooWorkspace::saveSnaphot(combined) replacing previous snapshot with name ModelConfig__snapshot
[#0] PROGRESS:Eval -- Running for SigXsecOverSM = 3
=== Using the following for ModelConfig ===
Observables: RooArgSet:: = (obs_x_channel1,weightVar,channelCat)
Parameters of Interest: RooArgSet:: = (SigXsecOverSM)
Nuisance Parameters: RooArgSet:: = (alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
Global Observables: RooArgSet:: = (nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
PDF: RooSimultaneous::simPdf[ indexCat=channelCat channel1=model_channel1 ] = 0.206686
Snapshot:
1) 0x49cb540 RooRealVar:: SigXsecOverSM = 3 L(0 - 3) "SigXsecOverSM"
=== Using the following for ModelConfig_with_poi_0 ===
Observables: RooArgSet:: = (obs_x_channel1,weightVar,channelCat)
Parameters of Interest: RooArgSet:: = (SigXsecOverSM)
Nuisance Parameters: RooArgSet:: = (alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
Global Observables: RooArgSet:: = (nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
PDF: RooSimultaneous::simPdf[ indexCat=channelCat channel1=model_channel1 ] = 0.206686
Snapshot:
1) 0x49cb540 RooRealVar:: SigXsecOverSM = 0 L(0 - 3) "SigXsecOverSM"
[#0] PROGRESS:Generation -- Test Statistic on data: 3.27476
[#1] INFO:InputArguments -- Profiling conditional MLEs for Null.
[#1] INFO:InputArguments -- Using a ToyMCSampler. Now configuring for Null.
[#0] PROGRESS:Generation -- generated toys: 500 / 1000
[#1] INFO:InputArguments -- Profiling conditional MLEs for Alt.
[#1] INFO:InputArguments -- Using a ToyMCSampler. Now configuring for Alt.
[#0] PROGRESS:Eval -- P values for SigXsecOverSM = 3
CLs = 0.00535332 +/- 0.00238893
CLb = 0.934 +/- 0.0111035
CLsplusb = 0.005 +/- 0.00223047
Time to perform limit scan
Real time 0:00:13, CP time 13.070
The computed upper limit is: 2.46135 +/- 0.0596845
Expected upper limits, using the B (alternate) model :
expected limit (median) 1.60988
expected limit (-1 sig) 1.34011
expected limit (+1 sig) 2.09019
expected limit (-2 sig) 1.14968
expected limit (+2 sig) 2.79445
[#0] WARNING:Plotting -- Could not determine xmin and xmax of sampling distribution that was given to plot.
[#0] WARNING:Plotting -- Could not determine xmin and xmax of sampling distribution that was given to plot.
#include <cassert>
struct HypoTestInvOptions {
bool plotHypoTestResult = true;
bool writeResult = true;
TString resultFileName;
bool optimize = true;
bool useVectorStore = true;
bool generateBinned = false;
bool noSystematics = false;
double nToysRatio = 2;
double maxPOI = -1;
bool useProof = false;
int nworkers = 0;
bool enableDetailedOutput = false;
bool rebuild = false;
int nToyToRebuild = 100;
int rebuildParamValues=0;
int initialFit = -1;
int randomSeed = -1;
int nAsimovBins = 0;
bool reuseAltToys = false;
double confLevel = 0.95;
std::string minimizerType = "";
std::string massValue = "";
int printLevel = 0;
bool useNLLOffset = false;
};
HypoTestInvOptions optHTInv;
class HypoTestInvTool{
public:
HypoTestInvTool();
~HypoTestInvTool(){};
const char * modelSBName, const char * modelBName,
const char * dataName,
int type, int testStatType,
bool useCLs,
int npoints, double poimin, double poimax, int ntoys,
bool useNumberCounting = false,
const char * nuisPriorName = 0);
void
int calculatorType,
int testStatType,
bool useCLs,
int npoints,
const char * fileNameBase = 0 );
void SetParameter(const char * name, const char * value);
void SetParameter(const char * name, bool value);
void SetParameter(const char * name, int value);
void SetParameter(const char * name, double value);
private:
bool mPlotHypoTestResult;
bool mWriteResult;
bool mOptimize;
bool mUseVectorStore;
bool mGenerateBinned;
bool mUseProof;
bool mRebuild;
bool mReuseAltToys;
bool mEnableDetOutput;
int mNWorkers;
int mNToyToRebuild;
int mRebuildParamValues;
int mPrintLevel;
int mInitialFit;
int mRandomSeed;
double mNToysRatio;
double mMaxPoi;
int mAsimovBins;
std::string mMassValue;
std::string mMinimizerType;
TString mResultFileName;
};
}
RooStats::HypoTestInvTool::HypoTestInvTool() : mPlotHypoTestResult(true),
mWriteResult(false),
mOptimize(true),
mUseVectorStore(true),
mGenerateBinned(false),
mUseProof(false),
mEnableDetOutput(false),
mRebuild(false),
mReuseAltToys(false),
mNWorkers(4),
mNToyToRebuild(100),
mRebuildParamValues(0),
mPrintLevel(0),
mInitialFit(-1),
mRandomSeed(-1),
mNToysRatio(2),
mMaxPoi(-1),
mAsimovBins(0),
mMassValue(""),
mMinimizerType(""),
mResultFileName() {
}
void
RooStats::HypoTestInvTool::SetParameter(const char * name, bool value){
std::string s_name(name);
if (s_name.find("PlotHypoTestResult") != std::string::npos) mPlotHypoTestResult = value;
if (s_name.find("WriteResult") != std::string::npos) mWriteResult = value;
if (s_name.find("Optimize") != std::string::npos) mOptimize = value;
if (s_name.find("UseVectorStore") != std::string::npos) mUseVectorStore = value;
if (s_name.find("GenerateBinned") != std::string::npos) mGenerateBinned = value;
if (s_name.find("UseProof") != std::string::npos) mUseProof = value;
if (s_name.find("EnableDetailedOutput") != std::string::npos) mEnableDetOutput = value;
if (s_name.find("Rebuild") != std::string::npos) mRebuild = value;
if (s_name.find("ReuseAltToys") != std::string::npos) mReuseAltToys = value;
return;
}
void
RooStats::HypoTestInvTool::SetParameter(const char * name, int value){
std::string s_name(name);
if (s_name.find("NWorkers") != std::string::npos) mNWorkers = value;
if (s_name.find("NToyToRebuild") != std::string::npos) mNToyToRebuild = value;
if (s_name.find("RebuildParamValues") != std::string::npos) mRebuildParamValues = value;
if (s_name.find("PrintLevel") != std::string::npos) mPrintLevel = value;
if (s_name.find("InitialFit") != std::string::npos) mInitialFit = value;
if (s_name.find("RandomSeed") != std::string::npos) mRandomSeed = value;
if (s_name.find("AsimovBins") != std::string::npos) mAsimovBins = value;
return;
}
void
RooStats::HypoTestInvTool::SetParameter(const char * name, double value){
std::string s_name(name);
if (s_name.find("NToysRatio") != std::string::npos) mNToysRatio = value;
if (s_name.find("MaxPOI") != std::string::npos) mMaxPoi = value;
return;
}
void
RooStats::HypoTestInvTool::SetParameter(const char * name, const char * value){
std::string s_name(name);
if (s_name.find("MassValue") != std::string::npos) mMassValue.assign(value);
if (s_name.find("MinimizerType") != std::string::npos) mMinimizerType.assign(value);
if (s_name.find("ResultFileName") != std::string::npos) mResultFileName = value;
return;
}
void
StandardHypoTestInvDemo(const char * infile = 0,
const char * wsName = "combined",
const char * modelSBName = "ModelConfig",
const char * modelBName = "",
const char * dataName = "obsData",
int calculatorType = 0,
int testStatType = 0,
bool useCLs = true ,
int npoints = 6,
double poimin = 0,
double poimax = 5,
int ntoys=1000,
bool useNumberCounting = false,
const char * nuisPriorName = 0){
TString filename(infile);
if (filename.IsNull()) {
filename = "results/example_combined_GaussExample_model.root";
if (!fileExist) {
#ifdef _WIN32
cout << "HistFactory file cannot be generated on Windows - exit" << endl;
return;
#endif
cout <<"will run standard hist2workspace example"<<endl;
gROOT->ProcessLine(
".! prepareHistFactory .");
gROOT->ProcessLine(
".! hist2workspace config/example.xml");
cout <<"\n\n---------------------"<<endl;
cout <<"Done creating example input"<<endl;
cout <<"---------------------\n\n"<<endl;
}
}
else
filename = infile;
if(!file ){
cout <<"StandardRooStatsDemoMacro: Input file " << filename << " is not found" << endl;
return;
}
HypoTestInvTool calc;
calc.SetParameter("PlotHypoTestResult", optHTInv.plotHypoTestResult);
calc.SetParameter("WriteResult", optHTInv.writeResult);
calc.SetParameter("Optimize", optHTInv.optimize);
calc.SetParameter("UseVectorStore", optHTInv.useVectorStore);
calc.SetParameter("GenerateBinned", optHTInv.generateBinned);
calc.SetParameter("NToysRatio", optHTInv.nToysRatio);
calc.SetParameter("MaxPOI", optHTInv.maxPOI);
calc.SetParameter("UseProof", optHTInv.useProof);
calc.SetParameter("EnableDetailedOutput", optHTInv.enableDetailedOutput);
calc.SetParameter("NWorkers", optHTInv.nworkers);
calc.SetParameter("Rebuild", optHTInv.rebuild);
calc.SetParameter("ReuseAltToys", optHTInv.reuseAltToys);
calc.SetParameter("NToyToRebuild", optHTInv.nToyToRebuild);
calc.SetParameter("RebuildParamValues", optHTInv.rebuildParamValues);
calc.SetParameter("MassValue", optHTInv.massValue.c_str());
calc.SetParameter("MinimizerType", optHTInv.minimizerType.c_str());
calc.SetParameter("PrintLevel", optHTInv.printLevel);
calc.SetParameter("InitialFit", optHTInv.initialFit);
calc.SetParameter("ResultFileName", optHTInv.resultFileName);
calc.SetParameter("RandomSeed", optHTInv.randomSeed);
calc.SetParameter("AsimovBins", optHTInv.nAsimovBins);
std::cout << w << "\t" << filename << std::endl;
if (w != NULL) {
r = calc.RunInverter(w, modelSBName, modelBName,
dataName, calculatorType, testStatType, useCLs,
npoints, poimin, poimax,
ntoys, useNumberCounting, nuisPriorName );
if (!r) {
std::cerr << "Error running the HypoTestInverter - Exit " << std::endl;
return;
}
}
else {
std::cout << "Reading an HypoTestInverterResult with name " << wsName << " from file " << filename << std::endl;
if (!r) {
std::cerr << "File " << filename << " does not contain a workspace or an HypoTestInverterResult - Exit "
<< std::endl;
return;
}
}
calc.AnalyzeResult( r, calculatorType, testStatType, useCLs, npoints, infile );
return;
}
void
int calculatorType,
int testStatType,
bool useCLs,
int npoints,
const char * fileNameBase ){
double lowerLimit = 0;
double llError = 0;
#if defined ROOT_SVN_VERSION && ROOT_SVN_VERSION >= 44126
}
#else
#endif
if (lowerLimit < upperLimit*(1.- 1.E-4) && lowerLimit != 0)
std::cout << "The computed lower limit is: " << lowerLimit << " +/- " << llError << std::endl;
std::cout << "The computed upper limit is: " << upperLimit << " +/- " << ulError << std::endl;
std::cout << "Expected upper limits, using the B (alternate) model : " << std::endl;
if (mEnableDetOutput) {
mWriteResult=true;
Info(
"StandardHypoTestInvDemo",
"detailed output will be written in output result file");
}
if (r != NULL && mWriteResult) {
const char * calcType = (calculatorType == 0) ? "Freq" : (calculatorType == 1) ? "Hybr" : "Asym";
const char * limitType = (useCLs) ? "CLs" : "Cls+b";
const char * scanType = (npoints < 0) ? "auto" : "grid";
if (mResultFileName.IsNull()) {
mResultFileName =
TString::Format(
"%s_%s_%s_ts%d_",calcType,limitType,scanType,testStatType);
if (mMassValue.size()>0) {
mResultFileName += mMassValue.c_str();
mResultFileName += "_";
}
TString name = fileNameBase;
name.Replace(0, name.Last('/')+1, "");
}
TString uldistFile = "RULDist.root";
if (existULDist) {
if (fileULDist) ulDist= fileULDist->Get("RULDist");
}
TFile * fileOut = new TFile(mResultFileName,"RECREATE");
if (ulDist) ulDist->
Write();
Info(
"StandardHypoTestInvDemo",
"HypoTestInverterResult has been written in the file %s",mResultFileName.Data());
fileOut->Close();
}
std::string typeName = "";
if (calculatorType == 0 )
typeName = "Frequentist";
if (calculatorType == 1 )
typeName = "Hybrid";
else if (calculatorType == 2 || calculatorType == 3) {
typeName = "Asymptotic";
mPlotHypoTestResult = false;
}
const char * resultName = r->
GetName();
TString plotTitle =
TString::Format(
"%s CL Scan for workspace %s",typeName.c_str(),resultName);
c1->SetLogy(false);
plot->Draw("CLb 2CL");
if (mPlotHypoTestResult) {
if (nEntries > 1) {
}
for (int i=0; i<nEntries; i++) {
if (nEntries > 1) c2->
cd(i+1);
}
}
}
const char * modelSBName, const char * modelBName,
const char * dataName, int type, int testStatType,
bool useCLs, int npoints, double poimin, double poimax,
int ntoys,
bool useNumberCounting,
const char * nuisPriorName ){
std::cout <<
"Running HypoTestInverter on the workspace " << w->
GetName() << std::endl;
if (!data) {
Error(
"StandardHypoTestDemo",
"Not existing data %s",dataName);
return 0;
}
else
std::cout << "Using data set " << dataName << std::endl;
if (mUseVectorStore) {
}
if (!sbModel) {
Error(
"StandardHypoTestDemo",
"Not existing ModelConfig %s",modelSBName);
return 0;
}
Error(
"StandardHypoTestDemo",
"Model %s has no pdf ",modelSBName);
return 0;
}
Error(
"StandardHypoTestDemo",
"Model %s has no poi ",modelSBName);
return 0;
}
Error(
"StandardHypoTestInvDemo",
"Model %s has no observables ",modelSBName);
return 0;
}
Info(
"StandardHypoTestInvDemo",
"Model %s has no snapshot - make one using model poi",modelSBName);
}
if (optHTInv.noSystematics) {
if (nuisPar && nuisPar->
getSize() > 0) {
std::cout << "StandardHypoTestInvDemo" << " - Switch off all systematics by setting them constant to their initial values" << std::endl;
}
if (bModel) {
if (bnuisPar)
}
}
if (!bModel || bModel == sbModel) {
Info(
"StandardHypoTestInvDemo",
"The background model %s does not exist",modelBName);
Info(
"StandardHypoTestInvDemo",
"Copy it from ModelConfig %s and set POI to zero",modelSBName);
bModel->
SetName(TString(modelSBName)+TString(
"_with_poi_0"));
if (!var) return 0;
double oldval = var->
getVal();
}
else {
Info(
"StandardHypoTestInvDemo",
"Model %s has no snapshot - make one using model poi and 0 values ",modelBName);
if (var) {
double oldval = var->
getVal();
}
else {
Error(
"StandardHypoTestInvDemo",
"Model %s has no valid poi",modelBName);
return 0;
}
}
}
if (type != 1 ) {
if (hasNuisParam && !hasGlobalObs ) {
if (constrPdf) {
Warning(
"StandardHypoTestInvDemo",
"Model %s has nuisance parameters but no global observables associated",sbModel->
GetName());
Warning(
"StandardHypoTestInvDemo",
"\tThe effect of the nuisance parameters will not be treated correctly ");
}
}
}
delete allParams;
std::cout <<
"StandardHypoTestInvDemo : POI initial value: " << poi->
GetName() <<
" = " << poi->
getVal() << std::endl;
bool doFit = mInitialFit;
if (testStatType == 0 && mInitialFit == -1) doFit = false;
if (type == 3 && mInitialFit == -1) doFit = false;
double poihat = 0;
else
Info(
"StandardHypoTestInvDemo",
"Using %s as minimizer for computing the test statistic",
if (doFit) {
Info(
"StandardHypoTestInvDemo",
" Doing a first fit to the observed data ");
Warning(
"StandardHypoTestInvDemo",
"Fit to the model failed - try with strategy 1 and perform first an Hesse computation");
}
Warning(
"StandardHypoTestInvDemo",
" Fit still failed - continue anyway.....");
std::cout <<
"StandardHypoTestInvDemo - Best Fit value : " << poi->
GetName() <<
" = " << poihat <<
" +/- " << poi->
getError() << std::endl;
std::cout <<
"Time for fitting : "; tw.
Print();
std::cout <<
"StandardHypoTestInvo: snapshot of S+B Model " << sbModel->
GetName()
<< " is set to the best fit value" << std::endl;
}
if (testStatType == 0) {
if (!doFit)
Info(
"StandardHypoTestInvDemo",
"Using LEP test statistic - an initial fit is not done and the TS will use the nuisances at the model value");
else
Info(
"StandardHypoTestInvDemo",
"Using LEP test statistic - an initial fit has been done and the TS will use the nuisances at the best fit value");
}
if (testStatType == 11) ropl.SetSubtractMLE(true);
ropl.SetPrintLevel(mPrintLevel);
ropl.SetMinimizer(mMinimizerType.c_str());
if (mEnableDetOutput) ropl.EnableDetailedOutput();
if (testStatType == 4) profll.
SetSigned(
true);
ropl.SetReuseNLL(mOptimize);
if (mOptimize) {
ropl.SetStrategy(0);
}
if (mMaxPoi > 0) poi->
setMax(mMaxPoi);
AsymptoticCalculator::SetPrintLevel(mPrintLevel);
else {
Error(
"StandardHypoTestInvDemo",
"Invalid - calculator type = %d supported values are only :\n\t\t\t 0 (Frequentist) , 1 (Hybrid) , 2 (Asymptotic) ",type);
return 0;
}
if (testStatType == 0) testStat = &slrts;
if (testStatType == 1 || testStatType == 11) testStat = &ropl;
if (testStatType == 2 || testStatType == 3 || testStatType == 4) testStat = &profll;
if (testStatType == 5) testStat = &maxll;
if (testStatType == 6) testStat = &nevtts;
if (testStat == 0) {
Error(
"StandardHypoTestInvDemo",
"Invalid - test statistic type = %d supported values are only :\n\t\t\t 0 (SLR) , 1 (Tevatron) , 2 (PLR), 3 (PLR1), 4(MLE)",testStatType);
return 0;
}
if (toymcs && (type == 0 || type == 1) ) {
if (useNumberCounting)
Warning(
"StandardHypoTestInvDemo",
"Pdf is extended: but number counting flag is set: ignore it ");
}
else {
if (!useNumberCounting ) {
Info(
"StandardHypoTestInvDemo",
"Pdf is not extended: number of events to generate taken from observed data set is %d",nEvents);
}
else {
Info(
"StandardHypoTestInvDemo",
"using a number counting pdf");
}
}
Info(
"StandardHypoTestInvDemo",
"Data set is weighted, nentries = %d and sum of weights = %8.1f but toy generation is unbinned - it would be faster to set mGenerateBinned to true\n",data->
numEntries(), data->
sumEntries());
}
Warning(
"StandardHypoTestInvDemo",
"generate binned is activated but the number of observable is %d. Too much memory could be needed for allocating all the bins",sbModel->
GetObservables()->
getSize() );
}
}
if (mReuseAltToys) {
}
if (type == 1) {
assert(hhc);
hhc->
SetToys(ntoys,ntoys/mNToysRatio);
ToyMCSampler::SetAlwaysUseMultiGen(false);
if (nuisPriorName) nuisPdf = w->
pdf(nuisPriorName);
if (!nuisPdf) {
Info(
"StandardHypoTestInvDemo",
"No nuisance pdf given for the HybridCalculator - try to deduce pdf from the model");
else
}
if (!nuisPdf ) {
Info(
"StandardHypoTestInvDemo",
"No nuisance pdf given - try to use %s that is defined as a prior pdf in the B model",nuisPdf->
GetName());
}
else {
Error(
"StandardHypoTestInvDemo",
"Cannot run Hybrid calculator because no prior on the nuisance parameter is specified or can be derived");
return 0;
}
}
assert(nuisPdf);
Info(
"StandardHypoTestInvDemo",
"Using as nuisance Pdf ... " );
Warning(
"StandardHypoTestInvDemo",
"Prior nuisance does not depend on nuisance parameters. They will be smeared in their full range");
}
delete np;
}
}
else if (type == 2 || type == 3) {
if (testStatType != 2 && testStatType != 3)
Warning(
"StandardHypoTestInvDemo",
"Only the PL test statistic can be used with AsymptoticCalculator - use by default a two-sided PL");
}
else if (type == 0 ) {
}
else if (type == 1 ) {
}
calc.SetConfidenceLevel(optHTInv.confLevel);
calc.UseCLs(useCLs);
calc.SetVerbose(true);
if (mUseProof) {
}
if (npoints > 0) {
if (poimin > poimax) {
poimin = int(poihat);
poimax = int(poihat + 4 * poi->
getError());
}
std::cout << "Doing a fixed scan in interval : " << poimin << " , " << poimax << std::endl;
calc.SetFixedScan(npoints,poimin,poimax);
}
else {
std::cout <<
"Doing an automatic scan in interval : " << poi->
getMin() <<
" , " << poi->
getMax() << std::endl;
}
std::cout << "Time to perform limit scan \n";
if (mRebuild) {
std::cout << "\n***************************************************************\n";
std::cout << "Rebuild the upper limit distribution by re-generating new set of pseudo-experiment and re-compute for each of them a new upper limit\n\n";
if (mRebuildParamValues != 0) {
*allParams = initialParameters;
}
if (mRebuildParamValues == 0 || mRebuildParamValues == 1 ) {
if (mRebuildParamValues == 0 ) {
std::cout << "rebuild using fitted parameter value for B-model snapshot" << std::endl;
constrainParams.
Print(
"v");
}
}
std::cout << "StandardHypoTestInvDemo: Initial parameters used for rebuilding: ";
delete allParams;
calc.SetCloseProof(1);
std::cout << "Time to rebuild distributions " << std::endl;
if (limDist) {
std::cout << "Expected limits after rebuild distribution " << std::endl;
std::cout <<
"expected upper limit (median of limit distribution) " << limDist->
InverseCDF(0.5) << std::endl;
limPlot.AddSamplingDistribution(limDist);
limPlot.GetTH1F()->SetStats(true);
limPlot.SetLineColor(kBlue);
new TCanvas(
"limPlot",
"Upper Limit Distribution");
limPlot.Draw();
TFile * fileOut = new TFile("RULDist.root","RECREATE");
fileOut->Close();
r = calc.GetInterval();
}
else
std::cout << "ERROR : failed to re-build distributions " << std::endl;
}
}
void ReadResult(const char * fileName, const char * resultName="", bool useCLs=true) {
StandardHypoTestInvDemo(fileName, resultName,"","","",0,0,useCLs);
}
#ifdef USE_AS_MAIN
StandardHypoTestInvDemo();
}
#endif



 Standard tutorial macro for performing an inverted hypothesis test for computing an interval
Standard tutorial macro for performing an inverted hypothesis test for computing an interval