
 Test program for the classes TUnfold and related.
Test program for the classes TUnfold and related.
Generate Monte Carlo and Data events The events consist of signal background
The signal is a resonance. It is generated with a Breit-Wigner, smeared by a Gaussian
Unfold the data. The result is: The background level The shape of the resonance, corrected for detector effects
Systematic errors from the MC shape variation are included and propagated to the result
- fit the unfolded distribution, including the correlation matrix
- save six plots to a file
testUnfold1.ps
- 2d-plot of the matrix describing the migrations
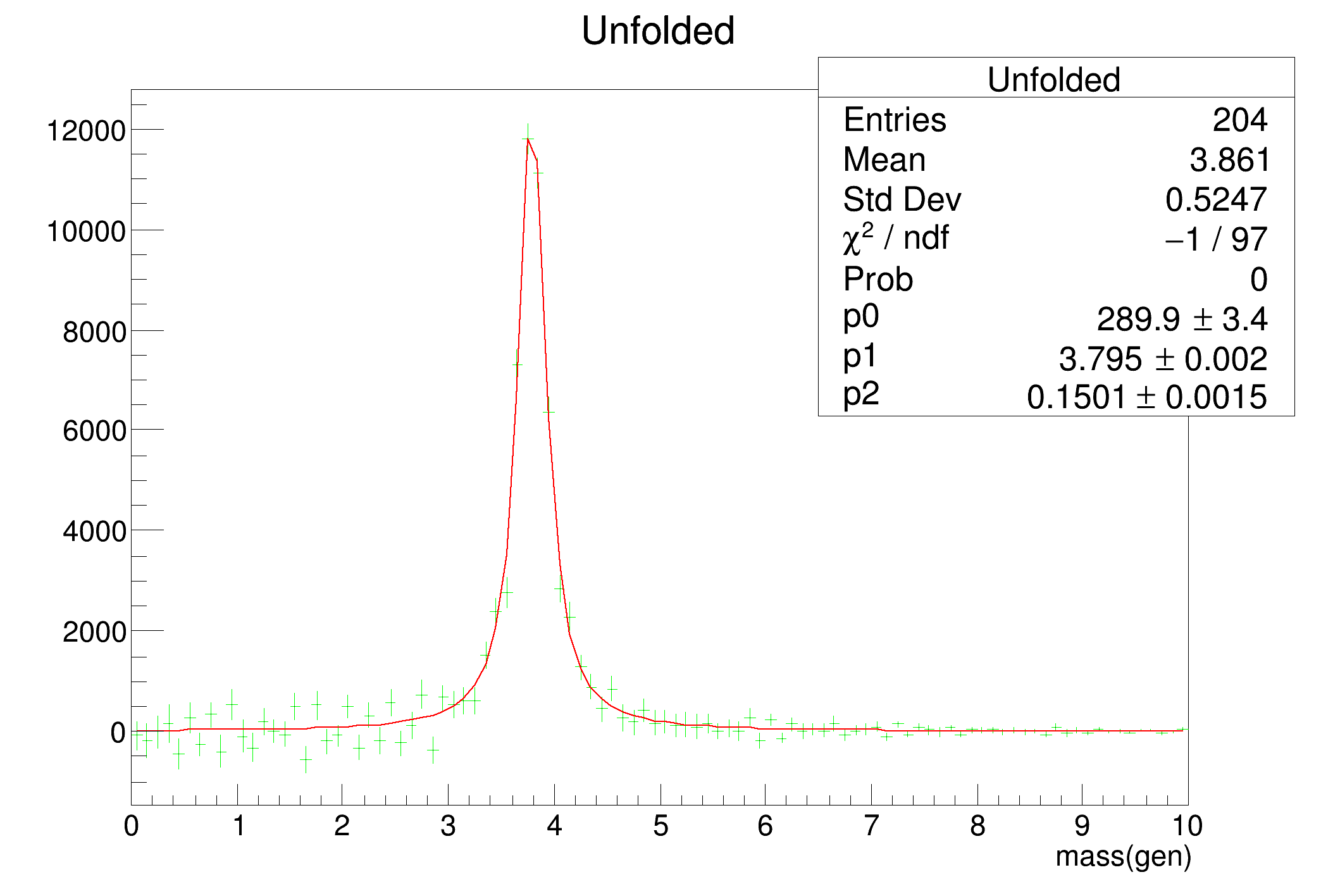
- generator-level distributions
- blue: unfolded data, total errors
- green: unfolded data, statistical errors
- red: generated data
- black: fit to green data points
- detector level distributions
- blue: unfolded data, folded back through the matrix
- black: Monte Carlo (with wrong peal position)
- blue: data
- global correlation coefficients
- \( \chi^2 \) as a function of \( log(\tau) \) the star indicates the final choice of \( \tau \)
- the L curve
0.0209980010986
7.82071614265
Processing /mnt/build/workspace/root-makedoc-v614/rootspi/rdoc/src/v6-14-00-patches/tutorials/unfold/testUnfold1.C...
tau=5.56618e-05
chi**2=173.079+9.63005 / 147
chi**2(sys)=147.813
FCN=99.8432 FROM MINOS STATUS=SUCCESSFUL 20 CALLS 190 TOTAL
EDM=4.83442e-07 STRATEGY= 1 ERROR MATRIX ACCURATE
EXT PARAMETER STEP FIRST
NO. NAME VALUE ERROR SIZE DERIVATIVE
1 p0 2.89860e+02 3.42457e+00 -1.98917e-03 -1.05299e-04
2 p1 3.79532e+00 1.74788e-03 1.06465e-06 -3.36308e-03
3 p2 1.50090e-01 1.46316e-03 1.46316e-03 -3.31677e-02
(int) 0
f = 0;
for (
Int_t i=0;i<nPoints;i++) {
else npfit++;
}
for (
Int_t i=0;i<nPoints;i++) {
for (
Int_t j=0;j<nPoints;j++) {
}
}
delete[] df;
}
return par[0]/(dm*dm+par[2]*par[2]);
}
{
do {
do {
} while(t>=1.0);
} while(t<=0.0);
return t;
} else {
do {
do {
} while(t>=1.0);
} while(t>=0.0);
return t;
}
}
return rnd->
Gaus(mTrue+smallBias,smallSigma);
} else {
return rnd->
Gaus(mTrue+wideBias,wideSigma);
}
}
int testUnfold1()
{
TH1D *histMgenMC=
new TH1D(
"MgenMC",
";mass(gen)",nGen,xminGen,xmaxGen);
TH1D *histMdetMC=
new TH1D(
"MdetMC",
";mass(det)",nDet,xminDet,xmaxDet);
TH2D *histMdetGenMC=
new TH2D(
"MdetgenMC",
";mass(det);mass(gen)",
nDet,xminDet,xmaxDet,nGen,xminGen,xmaxGen);
for(
Int_t i=0;i<neventMC;i++) {
4.0,
0.2);
histMgenMC->
Fill(mGen,luminosityData/luminosityMC);
histMdetMC->
Fill(mDet,luminosityData/luminosityMC);
histMdetGenMC->
Fill(mDet,mGen,luminosityData/luminosityMC);
}
TH2D *histMdetGenSysMC=
new TH2D(
"MdetgenSysMC",
";mass(det);mass(gen)",
nDet,xminDet,xmaxDet,nGen,xminGen,xmaxGen);
neventMC=rnd->
Poisson(luminosityMC*crossSection);
for(
Int_t i=0;i<neventMC;i++) {
(0.5,
3.6,
0.15);
histMdetGenSysMC->
Fill(mDet,mGen,luminosityData/luminosityMC);
}
TH1D *histMgenData=
new TH1D(
"MgenData",
";mass(gen)",nGen,xminGen,xmaxGen);
TH1D *histMdetData=
new TH1D(
"MdetData",
";mass(det)",nDet,xminDet,xmaxDet);
for(
Int_t i=0;i<neventData;i++) {
3.8,
0.15);
histMgenData->
Fill(mGen);
histMdetData->
Fill(mDet);
}
TH1D *histDensityGenData=
new TH1D(
"DensityGenData",
";mass(gen)",
nGen,xminGen,xmaxGen);
TH1D *histDensityGenMC=
new TH1D(
"DensityGenMC",
";mass(gen)",
nGen,xminGen,xmaxGen);
for(
Int_t i=1;i<=nGen;i++) {
}
if(unfold.SetInput(histMdetData)>=10000) {
std::cout<<"Unfolding result may be wrong\n";
}
#ifdef VERBOSE_LCURVE_SCAN
#endif
iBest=unfold.ScanLcurve(nScan,tauMin,tauMax,&lCurve,&logTauX,&logTauY);
#ifdef VERBOSE_LCURVE_SCAN
#endif
TH2D *histMdetGenSys1=
new TH2D(
"Mdetgensys1",
";mass(det);mass(gen)",
nDet,xminDet,xmaxDet,nGen,xminGen,xmaxGen);
for(
Int_t i=0;i<=nDet+1;i++) {
for(
Int_t j=0;j<=nGen+1;j++) {
}
}
}
std::cout<<"tau="<<unfold.GetTau()<<"\n";
std::cout<<"chi**2="<<unfold.GetChi2A()<<"+"<<unfold.GetChi2L()
<<" / "<<unfold.GetNdf()<<"\n";
std::cout<<"chi**2(sys)="<<unfold.GetChi2Sys()<<"\n";
TH1 *histMunfold=unfold.GetOutput(
"Unfolded");
TH1 *histMdetFold=unfold.GetFoldedOutput(
"FoldedBack");
TH2 *histEmatTotal=unfold.GetEmatrixTotal(
"EmatTotal");
new TH1D(
"TotalError",
";mass(gen)",nGen,xminGen,xmaxGen);
for(
Int_t bin=1;bin<=nGen;bin++) {
}
TH1 *histRhoi=unfold.GetRhoItotal(
"rho_I",
0,
0,
"*[UO]",
&gHistInvEMatrix
);
gFitter->
SetFCN(chisquare_corr);
TF1 *bw=
new TF1(
"bw",bw_func,xminGen,xmaxGen,3);
histMunfold->
Fit(bw,
"UE");
histMdetGenMC->
Draw(
"BOX");
histTotalError->
Draw(
"E");
histMunfold->
Draw(
"SAME E1");
histDensityGenData->
Draw(
"SAME");
histDensityGenMC->
Draw(
"SAME HIST");
histMdetMC->
Draw(
"SAME HIST");
TH1 *histInput=unfold.GetInput(
"Minput",
";mass(det)");
bestLogTauLogChi2->
Draw(
"*");
output.
SaveAs(
"testUnfold1.ps");
return 0;
}
Version 17.6, in parallel to changes in TUnfold
History:
- Version 17.5, in parallel to changes in TUnfold
- Version 17.4, in parallel to changes in TUnfold
- Version 17.3, in parallel to changes in TUnfold
- Version 17.2, in parallel to changes in TUnfold
- Version 17.1, in parallel to changes in TUnfold
- Version 17.0, updated for using the classes TUnfoldDensity, TUnfoldBinning
- Version 16.1, parallel to changes in TUnfold
- Version 16.0, parallel to changes in TUnfold
- Version 15, with automated L-curve scan
- Version 14, with changes in TUnfoldSys.cxx
- Version 13, include test of systematic errors
- Version 12, catch error when defining the input
- Version 11, print chi**2 and number of degrees of freedom
- Version 10, with bug-fix in TUnfold.cxx
- Version 9, with bug-fix in TUnfold.cxx and TUnfold.h
- Version 8, with bug-fix in TUnfold.cxx and TUnfold.h
- Version 7, with bug-fix in TUnfold.cxx and TUnfold.h
- Version 6a, fix problem with dynamic array allocation under windows
- Version 6, bug-fixes in TUnfold.C
- Version 5, replace main() by testUnfold1()
- Version 4, with bug-fix in TUnfold.C
- Version 3, with bug-fix in TUnfold.C
- Version 2, with changed ScanLcurve() arguments
- Version 1, remove L curve analysis, use ScanLcurve() method instead
- Version 0, L curve analysis included here
This file is part of TUnfold.
TUnfold is free software: you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation, either version 3 of the License, or (at your option) any later version.
TUnfold is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details.
You should have received a copy of the GNU General Public License along with TUnfold. If not, see http://www.gnu.org/licenses/.
- Author
- Stefan Schmitt DESY, 14.10.2008
Definition in file testUnfold1.C.



 Test program for the classes TUnfold and related.
Test program for the classes TUnfold and related. 