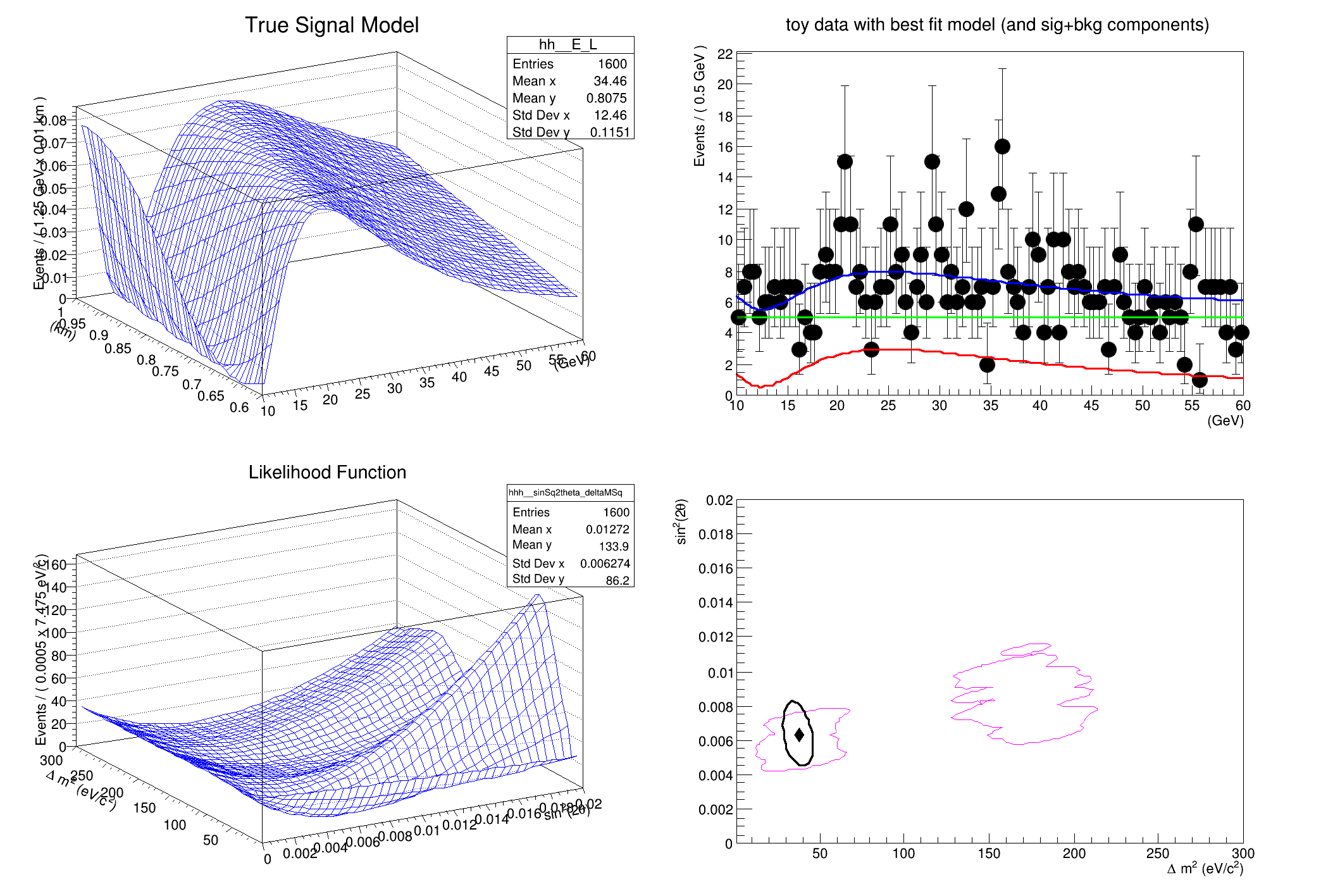
This tutorial shows a more complex example using the FeldmanCousins utility to create a confidence interval for a toy neutrino oscillation experiment. The example attempts to faithfully reproduce the toy example described in Feldman & Cousins' original paper, Phys.Rev.D57:3873-3889,1998.
␛[1mRooFit v3.60 -- Developed by Wouter Verkerke and David Kirkby␛[0m
Copyright (C) 2000-2013 NIKHEF, University of California & Stanford University
All rights reserved, please read http://roofit.sourceforge.net/license.txt
generate toy data with nEvents = 692
**********
** 1 **SET PRINT 1
**********
**********
** 2 **SET NOGRAD
**********
PARAMETER DEFINITIONS:
NO. NAME VALUE STEP SIZE LIMITS
1 deltaMSq 4.00000e+01 1.95000e+01 1.00000e+00 3.00000e+02
2 sinSq2theta 6.00000e-03 2.00000e-03 0.00000e+00 2.00000e-02
**********
** 3 **SET ERR 0.5
**********
**********
** 4 **SET PRINT 1
**********
**********
** 5 **SET STR 1
**********
NOW USING STRATEGY 1: TRY TO BALANCE SPEED AGAINST RELIABILITY
**********
** 6 **MIGRAD 1000 1
**********
FIRST CALL TO USER FUNCTION AT NEW START POINT, WITH IFLAG=4.
START MIGRAD MINIMIZATION. STRATEGY 1. CONVERGENCE WHEN EDM .LT. 1.00e-03
FCN=-1131.15 FROM MIGRAD STATUS=INITIATE 8 CALLS 9 TOTAL
EDM= unknown STRATEGY= 1 NO ERROR MATRIX
EXT PARAMETER CURRENT GUESS STEP FIRST
NO. NAME VALUE ERROR SIZE DERIVATIVE
1 deltaMSq 4.00000e+01 1.95000e+01 1.99953e-01 1.35503e+01
2 sinSq2theta 6.00000e-03 2.00000e-03 2.21072e-01 -1.80161e+00
ERR DEF= 0.5
MIGRAD MINIMIZATION HAS CONVERGED.
MIGRAD WILL VERIFY CONVERGENCE AND ERROR MATRIX.
COVARIANCE MATRIX CALCULATED SUCCESSFULLY
FCN=-1131.34 FROM MIGRAD STATUS=CONVERGED 32 CALLS 33 TOTAL
EDM=8.53321e-08 STRATEGY= 1 ERROR MATRIX ACCURATE
EXT PARAMETER STEP FIRST
NO. NAME VALUE ERROR SIZE DERIVATIVE
1 deltaMSq 3.75389e+01 4.12974e+00 9.32732e-04 7.25757e-03
2 sinSq2theta 6.29097e-03 8.61732e-04 2.04882e-03 6.82825e-04
ERR DEF= 0.5
EXTERNAL ERROR MATRIX. NDIM= 25 NPAR= 2 ERR DEF=0.5
1.706e+01 -1.140e-03
-1.140e-03 7.447e-07
PARAMETER CORRELATION COEFFICIENTS
NO. GLOBAL 1 2
1 0.31971 1.000 -0.320
2 0.31971 -0.320 1.000
**********
** 7 **SET ERR 0.5
**********
**********
** 8 **SET PRINT 1
**********
**********
** 9 **HESSE 1000
**********
COVARIANCE MATRIX CALCULATED SUCCESSFULLY
FCN=-1131.34 FROM HESSE STATUS=OK 10 CALLS 43 TOTAL
EDM=8.52801e-08 STRATEGY= 1 ERROR MATRIX ACCURATE
EXT PARAMETER INTERNAL INTERNAL
NO. NAME VALUE ERROR STEP SIZE VALUE
1 deltaMSq 3.75389e+01 4.12749e+00 3.73093e-05 -8.56559e-01
2 sinSq2theta 6.29097e-03 8.61259e-04 4.09765e-04 -3.79981e-01
ERR DEF= 0.5
EXTERNAL ERROR MATRIX. NDIM= 25 NPAR= 2 ERR DEF=0.5
1.705e+01 -1.133e-03
-1.133e-03 7.439e-07
PARAMETER CORRELATION COEFFICIENTS
NO. GLOBAL 1 2
1 0.31816 1.000 -0.318
2 0.31816 -0.318 1.000
[#1] INFO:Minization -- p.d.f. provides expected number of events, including extended term in likelihood.
[#1] INFO:NumericIntegration -- RooRealIntegral::init(PnmuTonePrime_Int[EPrime,LPrime]) using numeric integrator RooAdaptiveIntegratorND to calculate Int(LPrime,EPrime)
[#1] INFO:NumericIntegration -- RooRealIntegral::init(PnmuTone_Int[E,L]) using numeric integrator RooAdaptiveIntegratorND to calculate Int(L,E)
[#1] INFO:NumericIntegration -- RooRealIntegral::init(PnmuTone_Int[L]_Norm[E,L]) using numeric integrator RooIntegrator1D to calculate Int(L)
[#1] INFO:Minization -- createNLL picked up cached constraints from workspace with 0 entries
Metropolis-Hastings progress: ....................................................................................................
[#1] INFO:Eval -- Proposal acceptance rate: 3.3%
[#1] INFO:Eval -- Number of steps in chain: 165
[#1] INFO:NumericIntegration -- RooRealIntegral::init(product_Int[deltaMSq,sinSq2theta]_Norm[deltaMSq,sinSq2theta]) using numeric integrator RooAdaptiveIntegratorND to calculate Int(deltaMSq,sinSq2theta)
[#0] WARNING:NumericIntegration -- RooAdaptiveIntegratorND::dtor(product) WARNING: Number of suppressed warningings about integral evaluations where target precision was not reached is 1
[#1] INFO:NumericIntegration -- RooRealIntegral::init(product_Int[deltaMSq,sinSq2theta]_Norm[deltaMSq,sinSq2theta]) using numeric integrator RooAdaptiveIntegratorND to calculate Int(deltaMSq,sinSq2theta)
[#1] INFO:Eval -- cutoff = 0.166573, conf = 0.904333
[#0] WARNING:NumericIntegration -- RooAdaptiveIntegratorND::dtor(product) WARNING: Number of suppressed warningings about integral evaluations where target precision was not reached is 1
[#0] WARNING:NumericIntegration -- RooAdaptiveIntegratorND::dtor(PnmuTone) WARNING: Number of suppressed warningings about integral evaluations where target precision was not reached is 628
[#0] WARNING:NumericIntegration -- RooAdaptiveIntegratorND::dtor(PnmuTonePrime) WARNING: Number of suppressed warningings about integral evaluations where target precision was not reached is 628
Real time 0:02:38, CP time 158.690
MCMC actual confidence level: 0.904333
[#1] INFO:Minization -- RooProfileLL::evaluate(nll_model_modelData_Profile[deltaMSq,sinSq2theta]) Creating instance of MINUIT
[#1] INFO:Minization -- RooProfileLL::evaluate(nll_model_modelData_Profile[deltaMSq,sinSq2theta]) determining minimum likelihood for current configurations w.r.t all observable
[#1] INFO:Minization -- RooProfileLL::evaluate(nll_model_modelData_Profile[deltaMSq,sinSq2theta]) minimum found at (deltaMSq=37.5376, sinSq2theta=0.00629099)
..[#1] INFO:Minization -- LikelihoodInterval - Finding the contour of deltaMSq ( 0 ) and sinSq2theta ( 1 )
Real time 0:03:16, CP time 196.280
#include <iostream>
#if !defined(__CINT__) || defined(__MAKECINT__)
#include "../tutorials/roostats/NuMuToNuE_Oscillation.h"
#include "../tutorials/roostats/NuMuToNuE_Oscillation.cxx"
#else
#include "../tutorials/roostats/NuMuToNuE_Oscillation.cxx+"
#endif
void rs401d_FeldmanCousins(bool doFeldmanCousins = false, bool doMCMC = true)
{
RooRealVar deltaMSq(
"deltaMSq",
"#Delta m^{2}", 40, 1, 300,
"eV/c^{2}");
RooRealVar sinSq2theta(
"sinSq2theta",
"sin^{2}(2#theta)", .006, .0, .02);
RooRealVar EPrime(
"EPrime",
"", 15, 10, 60,
"GeV");
RooRealVar LPrime(
"LPrime",
"", .800, .600, 1.0,
"km");
NuMuToNuE_Oscillation PnmuTonePrime(
"PnmuTonePrime",
"P(#nu_{#mu} #rightarrow #nu_{e}", LPrime, EPrime, deltaMSq);
RooConstVar maxEventsTot(
"maxEventsTot",
"maximum number of sinal events", 50000);
RooConstVar inverseArea(
"inverseArea",
"1/(#Delta E #Delta L)",
1. / (EPrime.getMax() - EPrime.getMin()) / (LPrime.getMax() - LPrime.getMin()));
RooProduct sigNorm(
"sigNorm",
"",
RooArgSet(maxEventsTot, *intProbToOscInExp, inverseArea, sinSq2theta));
RooConstVar bkgNorm(
"bkgNorm",
"normalization for background", 500);
Int_t nEventsData = bkgNorm.getVal() + sigNorm.getVal();
cout << "generate toy data with nEvents = " << nEventsData << endl;
TH1 *hh = PnmuTone.createHistogram(
"hh", E, Binning(40), YVar(L, Binning(40)), Scaling(
kFALSE));
model.fitTo(*data, Extended());
model.plotOn(Eframe);
model.plotOn(Eframe, Components(*sigModel), LineColor(
kRed));
model.plotOn(Eframe, Components(bkgEShape), LineColor(
kGreen));
model.plotOn(Eframe);
Eframe->
SetTitle(
"toy data with best fit model (and sig+bkg components)");
RooNLLVar nll(
"nll",
"nll", model, *data, Extended());
TH1 *hhh = pll.createHistogram(
"hhh", sinSq2theta, Binning(40), YVar(deltaMSq, Binning(40)), Scaling(
kFALSE));
fc.UseAdaptiveSampling(
true);
if (doFeldmanCousins)
interval =
fc.GetInterval();
plc.SetTestSize(.1);
if (doMCMC) {
mc.SetNumIters(5000);
mc.SetNumBurnInSteps(100);
mc.SetUseKeys(true);
mc.SetTestSize(.1);
mc.SetAxes(axisList);
}
if (doFeldmanCousins) {
if (interval) {
} else {
}
}
delete tmpPoint;
}
if (interval) {
forContour->
Draw(
"cont2,same");
}
}
if (mcInt) {
}
plotInt.SetTitle("90% Confidence Intervals");
if (mcInt)
plotInt.Draw("same");
else
plotInt.Draw();
}
static struct mg_connection * fc(struct mg_context *ctx)
Double_t getRealValue(const char *name, Double_t defVal=0, Bool_t verbose=kFALSE) const
Get value of a RooAbsReal stored in set with given name.
TH1 * createHistogram(const char *name, const RooAbsRealLValue &xvar, const RooCmdArg &arg1=RooCmdArg::none(), const RooCmdArg &arg2=RooCmdArg::none(), const RooCmdArg &arg3=RooCmdArg::none(), const RooCmdArg &arg4=RooCmdArg::none(), const RooCmdArg &arg5=RooCmdArg::none(), const RooCmdArg &arg6=RooCmdArg::none(), const RooCmdArg &arg7=RooCmdArg::none(), const RooCmdArg &arg8=RooCmdArg::none()) const
Calls createHistogram(const char *name, const RooAbsRealLValue& xvar, const RooLinkedList& argList) c...
virtual RooPlot * plotOn(RooPlot *frame, const RooCmdArg &arg1=RooCmdArg::none(), const RooCmdArg &arg2=RooCmdArg::none(), const RooCmdArg &arg3=RooCmdArg::none(), const RooCmdArg &arg4=RooCmdArg::none(), const RooCmdArg &arg5=RooCmdArg::none(), const RooCmdArg &arg6=RooCmdArg::none(), const RooCmdArg &arg7=RooCmdArg::none(), const RooCmdArg &arg8=RooCmdArg::none()) const
virtual RooAbsPdf * createProjection(const RooArgSet &iset)
Return a p.d.f that represent a projection of this p.d.f integrated over given observables.
RooAbsReal is the common abstract base class for objects that represent a real value and implements f...
RooAbsReal * createIntegral(const RooArgSet &iset, const RooCmdArg &arg1, const RooCmdArg &arg2=RooCmdArg::none(), const RooCmdArg &arg3=RooCmdArg::none(), const RooCmdArg &arg4=RooCmdArg::none(), const RooCmdArg &arg5=RooCmdArg::none(), const RooCmdArg &arg6=RooCmdArg::none(), const RooCmdArg &arg7=RooCmdArg::none(), const RooCmdArg &arg8=RooCmdArg::none()) const
Create an object that represents the integral of the function over one or more observables listed in ...
RooAddPdf is an efficient implementation of a sum of PDFs of the form.
RooArgList is a container object that can hold multiple RooAbsArg objects.
RooArgSet is a container object that can hold multiple RooAbsArg objects.
TObject * clone(const char *newname) const override
RooConstVar represent a constant real-valued object.
The RooDataHist is a container class to hold N-dimensional binned data.
Int_t numEntries() const override
Return the number of bins.
const RooArgSet * get() const override
Get bin centre of current bin.
RooDataSet is a container class to hold unbinned data.
void setStreamStatus(Int_t id, Bool_t active)
(De)Activate stream with given unique ID
static RooMsgService & instance()
Return reference to singleton instance.
Class RooNLLVar implements a -log(likelihood) calculation from a dataset and a PDF.
A RooPlot is a plot frame and a container for graphics objects within that frame.
void SetTitle(const char *name)
Set the title of the RooPlot to 'title'.
virtual void Draw(Option_t *options=0)
Draw this plot and all of the elements it contains.
RooPolynomial implements a polynomial p.d.f of the form.
A RooProduct represents the product of a given set of RooAbsReal objects.
Class RooProfileLL implements the profile likelihood estimator for a given likelihood and set of para...
static TRandom * randomGenerator()
Return a pointer to a singleton random-number generator implementation.
RooRealVar represents a variable that can be changed from the outside.
ConfInterval is an interface class for a generic interval in the RooStats framework.
virtual Bool_t IsInInterval(const RooArgSet &) const =0
check if given point is in the interval
The FeldmanCousins class (like the Feldman-Cousins technique) is essentially a specific configuration...
This class provides simple and straightforward utilities to plot a LikelihoodInterval object.
LikelihoodInterval is a concrete implementation of the RooStats::ConfInterval interface.
Bayesian Calculator estimating an interval or a credible region using the Markov-Chain Monte Carlo me...
This class provides simple and straightforward utilities to plot a MCMCInterval object.
void SetLineColor(Color_t color)
void Draw(const Option_t *options=NULL)
MCMCInterval is a concrete implementation of the RooStats::ConfInterval interface.
virtual Double_t GetActualConfidenceLevel()
virtual Double_t GetKeysPdfCutoff() { return fKeysCutoff; }
ModelConfig is a simple class that holds configuration information specifying how a model should be u...
virtual void SetWorkspace(RooWorkspace &ws)
virtual void SetParametersOfInterest(const RooArgSet &set)
Specify parameters of interest.
virtual void SetPdf(const RooAbsPdf &pdf)
Set the Pdf, add to the the workspace if not already there.
The ProfileLikelihoodCalculator is a concrete implementation of CombinedCalculator (the interface cla...
The RooWorkspace is a persistable container for RooFit projects.
virtual void SetLineWidth(Width_t lwidth)
Set the line width.
virtual void SetLineColor(Color_t lcolor)
Set the line color.
TVirtualPad * cd(Int_t subpadnumber=0) override
Set current canvas & pad.
void Update() override
Update canvas pad buffers.
TH1 is the base class of all histogram classes in ROOT.
virtual void SetTitle(const char *title)
See GetStatOverflows for more information.
TObject * Clone(const char *newname=0) const
Make a complete copy of the underlying object.
virtual void SetContour(Int_t nlevels, const Double_t *levels=0)
Set the number and values of contour levels.
virtual void Draw(Option_t *option="")
Draw this histogram with options.
virtual Int_t FindBin(Double_t x, Double_t y=0, Double_t z=0)
Return Global bin number corresponding to x,y,z.
2-D histogram with a float per channel (see TH1 documentation)}
virtual void SetBinContent(Int_t bin, Double_t content)
Set bin content.
void Divide(Int_t nx=1, Int_t ny=1, Float_t xmargin=0.01, Float_t ymargin=0.01, Int_t color=0) override
Automatic pad generation by division.
virtual void SetSeed(ULong_t seed=0)
Set the random generator seed.
void Start(Bool_t reset=kTRUE)
Start the stopwatch.
void Stop()
Stop the stopwatch.
void Print(Option_t *option="") const
Print the real and cpu time passed between the start and stop events.
The namespace RooFit contains mostly switches that change the behaviour of functions of PDFs (or othe...
Namespace for the RooStats classes.

 Neutrino Oscillation Example from Feldman & Cousins
Neutrino Oscillation Example from Feldman & Cousins