

OneSidedFrequentistUpperLimitWithBands
This is a standard demo that can be used with any ROOT file prepared in the standard way. You specify:
- name for input ROOT file
- name of workspace inside ROOT file that holds model and data
- name of ModelConfig that specifies details for calculator tools
- name of dataset
With default parameters the macro will attempt to run the standard hist2workspace example and read the ROOT file that it produces.
The first ~100 lines define a new test statistic, then the main macro starts. You may want to control:
double confidenceLevel=0.95;
int nPointsToScan = 12;
int nToyMC = 150;
This uses a modified version of the profile likelihood ratio as a test statistic for upper limits (eg. test stat = 0 if muhat>mu).
Based on the observed data, one defines a set of parameter points to be tested based on the value of the parameter of interest and the conditional MLE (eg. profiled) values of the nuisance parameters.
At each parameter point, pseudo-experiments are generated using this fixed reference model and then the test statistic is evaluated. Note, the nuisance parameters are floating in the fits. For each point, the threshold that defines the 95% acceptance region is found. This forms a "Confidence Belt".
After constructing the confidence belt, one can find the confidence interval for any particular dataset by finding the intersection of the observed test statistic and the confidence belt. First this is done on the observed data to get an observed 1-sided upper limt.
Finally, there expected limit and bands (from background-only) are formed by generating background-only data and finding the upper limit. This is done by hand for now, will later be part of the RooStats tools.
On a technical note, this technique is NOT the Feldman-Cousins technique, because that is a 2-sided interval BY DEFINITION. However, like the Feldman-Cousins technique this is a Neyman-Construction. For technical reasons the easiest way to implement this right now is to use the FeldmanCousins tool and then change the test statistic that it is using.
Building the confidence belt can be computationally expensive. Once it is built, one could save it to a file and use it in a separate step.
We can use PROOF to speed things along in parallel, however, the test statistic has to be installed on the workers so either turn off PROOF or include the modified test statistic in your $ROOTSYS/roofit/roostats/inc directory, add the additional line to the LinkDef.h file, and recompile root.
Note, if you have a boundary on the parameter of interest (eg. cross-section) the threshold on the one-sided test statistic starts off very small because we are only including downward fluctuations. You can see the threshold in these printouts:
[#0] PROGRESS:Generation -- generated toys: 500 / 999
NeymanConstruction: Prog: 12/50
total MC = 39
this test stat = 0
SigXsecOverSM=0.69 alpha_syst1=0.136515 alpha_syst3=0.425415 beta_syst2=1.08496 [-1
e+30, 0.011215] in interval = 1
static unsigned int total
this tells you the values of the parameters being used to generate the pseudo-experiments and the threshold in this case is 0.011215. One would expect for 95% that the threshold would be ~1.35 once the cross-section is far enough away from 0 that it is essentially unaffected by the boundary. As one reaches the last points in the scan, the theshold starts to get artificially high. This is because the range of the parameter in the fit is the same as the range in the scan. In the future, these should be independently controlled, but they are not now. As a result the ~50% of pseudo-experiments that have an upward fluctuation end up with muhat = muMax. Because of this, the upper range of the parameter should be well above the expected upper limit... but not too high or one will need a very large value of nPointsToScan to resolve the relevant region. This can be improved, but this is the first version of this script.
Important note: when the model includes external constraint terms, like a Gaussian constraint to a nuisance parameter centered around some nominal value there is a subtlety. The asymptotic results are all based on the assumption that all the measurements fluctuate... including the nominal values from auxiliary measurements. If these do not fluctuate, this corresponds to an "conditional ensemble". The result is that the distribution of the test statistic can become very non-chi^2. This results in thresholds that become very large. This can be seen in the following thought experiment. Say the model is \( Pois(N | s + b)G(b0|b,sigma) \) where \( G(b0|b,sigma) \) is the external constraint and b0 is 100. If N is also 100 then the profiled value of b given s is going to be some trade off between 100-s and b0. If sigma is \( \sqrt(N) \), then the profiled value of b is probably 100 - s/2 So for s=60 we are going to have a profiled value of b~70. Now when we generate pseudo-experiments for s=60, b=70 we will have N~130 and the average shat will be 30, not 60. In practice, this is only an issue for values of s that are very excluded. For values of s near the 95% limit this should not be a big effect. This can be avoided if the nominal values of the constraints also fluctuate, but that requires that those parameters are RooRealVars in the model. This version does not deal with this issue, but it will be addressed in a future version.
FeldmanCousins: ntoys per point = 499
FeldmanCousins: nEvents per toy will fluctuate about expectation
will use global observables for unconditional ensemble
RooArgSet:: = (nominalLumi,nom_alpha_syst1,nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
=== Using the following for ModelConfig ===
Observables: RooArgSet:: = (obs_x_channel1,channelCat)
Parameters of Interest: RooArgSet:: = (SigXsecOverSM)
Nuisance Parameters: RooArgSet:: = (alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
Global Observables: RooArgSet:: = (nominalLumi,nom_alpha_syst1,nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
PDF: RooSimultaneous::simPdf[ indexCat=channelCat channel1=model_channel1 ] = 0.190787
FeldmanCousins: Model has nuisance parameters, will do profile construction
FeldmanCousins: # points to test = 12
lookup index = 0
NeymanConstruction: Prog: 1/12 total MC = 499 this test stat = 0
SigXsecOverSM=0.125 alpha_syst2=0.619973 alpha_syst3=0.233361 gamma_stat_channel1_bin_0=1.03213 gamma_stat_channel1_bin_1=1.04741 [-1e+30, 0.329527] in interval = 1
NeymanConstruction: Prog: 2/12 total MC = 499 this test stat = 0
SigXsecOverSM=0.375 alpha_syst2=0.44772 alpha_syst3=0.177828 gamma_stat_channel1_bin_0=1.02318 gamma_stat_channel1_bin_1=1.03602 [-1e+30, 0.886709] in interval = 1
NeymanConstruction: Prog: 3/12 total MC = 499 this test stat = 0
SigXsecOverSM=0.625 alpha_syst2=0.286415 alpha_syst3=0.123093 gamma_stat_channel1_bin_0=1.01471 gamma_stat_channel1_bin_1=1.02485 [-1e+30, 1.23248] in interval = 1
NeymanConstruction: Prog: 4/12 total MC = 499 this test stat = 0
SigXsecOverSM=0.875 alpha_syst2=0.135426 alpha_syst3=0.0710021 gamma_stat_channel1_bin_0=1.00682 gamma_stat_channel1_bin_1=1.0134 [-1e+30, 1.51788] in interval = 1
NeymanConstruction: Prog: 5/12 total MC = 499 this test stat = 0.000123945
SigXsecOverSM=1.125 alpha_syst2=-0.0142596 alpha_syst3=0.0160367 gamma_stat_channel1_bin_0=0.999288 gamma_stat_channel1_bin_1=1.00312 [-1e+30, 1.54201] in interval = 1
NeymanConstruction: Prog: 6/12 total MC = 499 this test stat = 0.0914825
SigXsecOverSM=1.375 alpha_syst2=-0.157842 alpha_syst3=-0.0395143 gamma_stat_channel1_bin_0=0.992184 gamma_stat_channel1_bin_1=0.993117 [-1e+30, 1.76272] in interval = 1
NeymanConstruction: Prog: 7/12 total MC = 499 this test stat = 0.348977
SigXsecOverSM=1.625 alpha_syst2=-0.293106 alpha_syst3=-0.0887909 gamma_stat_channel1_bin_0=0.985748 gamma_stat_channel1_bin_1=0.98241 [-1e+30, 1.57284] in interval = 1
NeymanConstruction: Prog: 8/12 total MC = 499 this test stat = 0.767852
SigXsecOverSM=1.875 alpha_syst2=-0.422663 alpha_syst3=-0.140541 gamma_stat_channel1_bin_0=0.979593 gamma_stat_channel1_bin_1=0.972417 [-1e+30, 1.52915] in interval = 1
NeymanConstruction: Prog: 9/12 total MC = 499 this test stat = 1.34349
SigXsecOverSM=2.125 alpha_syst2=-0.544235 alpha_syst3=-0.191156 gamma_stat_channel1_bin_0=0.973827 gamma_stat_channel1_bin_1=0.96257 [-1e+30, 1.20669] in interval = 0
NeymanConstruction: Prog: 10/12 total MC = 499 this test stat = 2.07144
SigXsecOverSM=2.375 alpha_syst2=-0.657512 alpha_syst3=-0.240962 gamma_stat_channel1_bin_0=0.968396 gamma_stat_channel1_bin_1=0.952936 [-1e+30, 1.85229] in interval = 0
NeymanConstruction: Prog: 11/12 total MC = 499 this test stat = 2.94737
SigXsecOverSM=2.625 alpha_syst2=-0.763058 alpha_syst3=-0.290526 gamma_stat_channel1_bin_0=0.963223 gamma_stat_channel1_bin_1=0.943652 [-1e+30, 1.1503] in interval = 0
NeymanConstruction: Prog: 12/12 total MC = 499 this test stat = 3.9668
SigXsecOverSM=2.875 alpha_syst2=-0.861405 alpha_syst3=-0.338707 gamma_stat_channel1_bin_0=0.958362 gamma_stat_channel1_bin_1=0.934519 [-1e+30, 1.2709] in interval = 0
[#1] INFO:Eval -- 8 points in interval
95% interval on SigXsecOverSM is : [0.125, 1.875]
[#1] INFO:Minimization -- p.d.f. provides expected number of events, including extended term in likelihood.
[#1] INFO:Minimization -- Including the following constraint terms in minimization: (lumiConstraint,alpha_syst1Constraint,alpha_syst2Constraint,alpha_syst3Constraint,gamma_stat_channel1_bin_0_constraint,gamma_stat_channel1_bin_1_constraint)
[#1] INFO:Minimization -- The global observables are not defined , normalize constraints with respect to the parameters (Lumi,SigXsecOverSM,alpha_syst1,alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
[#1] INFO:Minimization -- RooProfileLL::evaluate(nll_simPdf_obsData_with_constr_Profile[SigXsecOverSM]) Creating instance of MINUIT
[#1] INFO:Fitting -- RooAddition::defaultErrorLevel(nll_simPdf_obsData_with_constr) Summation contains a RooNLLVar, using its error level
[#1] INFO:Minimization -- RooProfileLL::evaluate(nll_simPdf_obsData_with_constr_Profile[SigXsecOverSM]) determining minimum likelihood for current configurations w.r.t all observable
RooAbsTestStatistic::initSimMode: creating slave calculator #0 for state channel1 (2 dataset entries)
[#1] INFO:Fitting -- RooAbsTestStatistic::initSimMode: created 1 slave calculators.
[#1] INFO:Minimization -- RooProfileLL::evaluate(nll_simPdf_obsData_with_constr_Profile[SigXsecOverSM]) minimum found at (SigXsecOverSM=1.11589)
.
Will use these parameter points to generate pseudo data for bkg only
1) 0x55cde64b6fa0 RooRealVar:: alpha_syst2 = 0.711146 +/- 0.91413 L(-5 - 5) "alpha_syst2"
2) 0x55cde64684a0 RooRealVar:: alpha_syst3 = 0.261455 +/- 0.929175 L(-5 - 5) "alpha_syst3"
3) 0x55cde6471ce0 RooRealVar:: gamma_stat_channel1_bin_0 = 1.03677 +/- 0.0462907 L(0 - 1.25) "gamma_stat_channel1_bin_0"
4) 0x55cde643f7f0 RooRealVar:: gamma_stat_channel1_bin_1 = 1.05319 +/- 0.0761269 L(0 - 1.5) "gamma_stat_channel1_bin_1"
5) 0x55cde6441840 RooRealVar:: SigXsecOverSM = 0 +/- 0 L(0 - 3) B(12) "SigXsecOverSM"
-2 sigma band 4.94066e-324
-1 sigma band 0.105 [Power Constraint)]
median of band 0.855
+1 sigma band 1.605
+2 sigma band 2.085
observed 95% upper-limit 1.875
CLb strict [P(toy>obs|0)] for observed 95% upper-limit 0.966667
CLb inclusive [P(toy>=obs|0)] for observed 95% upper-limit 0.966667
bool useProof = false;
int nworkers = 0;
void OneSidedFrequentistUpperLimitWithBands(const char *infile = "", const char *workspaceName = "combined",
const char *modelConfigName = "ModelConfig",
const char *dataName = "obsData")
{
double confidenceLevel = 0.95;
int nPointsToScan = 12;
int nToyMC = 150;
if (!strcmp(infile, "")) {
filename =
"results/example_combined_GaussExample_model.root";
if (!fileExist) {
#ifdef _WIN32
cout << "HistFactory file cannot be generated on Windows - exit" << endl;
return;
#endif
cout << "will run standard hist2workspace example" << endl;
gROOT->ProcessLine(
".! prepareHistFactory .");
gROOT->ProcessLine(
".! hist2workspace config/example.xml");
cout << "\n\n---------------------" << endl;
cout << "Done creating example input" << endl;
cout << "---------------------\n\n" << endl;
}
} else
cout <<
"StandardRooStatsDemoMacro: Input file " <<
filename <<
" is not found" << endl;
return;
}
cout << "workspace not found" << endl;
return;
}
cout << "data or ModelConfig was not found" << endl;
return;
}
fc.SetConfidenceLevel(confidenceLevel);
fc.AdditionalNToysFactor(
0.5);
fc.SetNBins(nPointsToScan);
fc.CreateConfBelt(true);
if (
data->numEntries() == 1)
fc.FluctuateNumDataEntries(false);
else
cout << "Not sure what to do about this model" << endl;
}
if (useProof) {
}
cout << "will use global observables for unconditional ensemble" << endl;
}
cout <<
"\n95% interval on " << firstPOI->
GetName() <<
" is : [" << interval->
LowerLimit(*firstPOI) <<
", " << interval->
UpperLimit(*firstPOI) <<
"] " << endl;
double observedUL = interval->
UpperLimit(*firstPOI);
double obsTSatObsUL = fc.GetTestStatSampler()->EvaluateTestStatistic(*
data, tmpPOI);
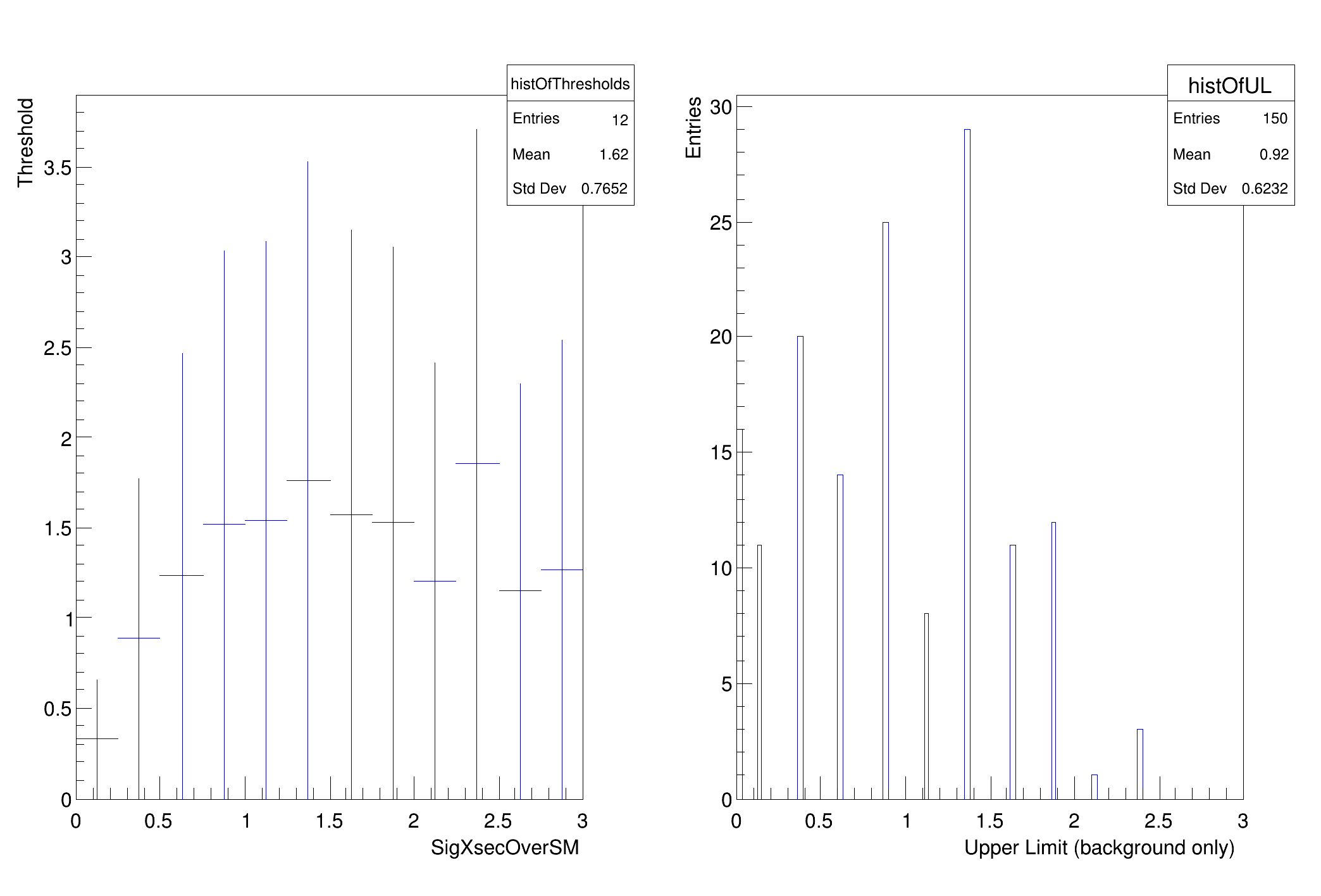
histOfThresholds->
Fill(poiVal, arMax);
}
histOfThresholds->
Draw();
profile->getVal();
w->saveSnapshot(
"paramsToGenerateData", *poiAndNuisance);
cout << "\nWill use these parameter points to generate pseudo data for bkg only" << endl;
paramsToGenerateData->
Print(
"v");
double CLb = 0;
double CLbinclusive = 0;
TH1F *histOfUL =
new TH1F(
"histOfUL",
"", 100, 0, firstPOI->
getMax());
for (int imc = 0; imc < nToyMC; ++imc) {
w->loadSnapshot(
"paramsToGenerateData");
if (
data->numEntries() == 1)
else
cout << "Not sure what to do about this model" << endl;
} else {
}
if (!simPdf) {
allVars->assign(*values);
delete values;
delete one;
} else {
for (
auto const&
tt : simPdf->indexCat()) {
auto const& catName =
tt.first;
std::unique_ptr<RooDataSet> tmp{pdftmp->
generate(*globtmp, 1)};
globtmp->assign(*tmp->get(0));
}
}
double toyTSatObsUL = fc.GetTestStatSampler()->EvaluateTestStatistic(*toyData, tmpPOI);
if (obsTSatObsUL < toyTSatObsUL)
CLb += (1.) / nToyMC;
if (obsTSatObsUL <= toyTSatObsUL)
CLbinclusive += (1.) / nToyMC;
double thisUL = 0;
double thisTS = fc.GetTestStatSampler()->EvaluateTestStatistic(*toyData, tmpPOI);
if (thisTS <= arMax) {
} else {
break;
}
}
delete toyData;
}
c1->SaveAs(
"one-sided_upper_limit_output.pdf");
double band2sigDown, band1sigDown, bandMedian, band1sigUp, band2sigUp;
for (
int i = 1; i <= cumulative->
GetNbinsX(); ++i) {
if (bins[i] < 0.5)
}
cout << "-2 sigma band " << band2sigDown << endl;
cout << "-1 sigma band " << band1sigDown << " [Power Constraint)]" << endl;
cout << "median of band " << bandMedian << endl;
cout << "+1 sigma band " << band1sigUp << endl;
cout << "+2 sigma band " << band2sigUp << endl;
cout <<
"\nobserved 95% upper-limit " << interval->
UpperLimit(*firstPOI) << endl;
cout << "CLb strict [P(toy>obs|0)] for observed 95% upper-limit " << CLb << endl;
cout << "CLb inclusive [P(toy>=obs|0)] for observed 95% upper-limit " << CLbinclusive << endl;
}
Option_t Option_t TPoint TPoint const char GetTextMagnitude GetFillStyle GetLineColor GetLineWidth GetMarkerStyle GetTextAlign GetTextColor GetTextSize void data
Option_t Option_t TPoint TPoint const char GetTextMagnitude GetFillStyle GetLineColor GetLineWidth GetMarkerStyle GetTextAlign GetTextColor GetTextSize void char Point_t Rectangle_t WindowAttributes_t Float_t Float_t Float_t Int_t Int_t UInt_t UInt_t Rectangle_t Int_t Int_t Window_t TString Int_t GCValues_t GetPrimarySelectionOwner GetDisplay GetScreen GetColormap GetNativeEvent const char const char dpyName wid window const char font_name cursor keysym reg const char only_if_exist regb h Point_t winding char text const char depth char const char Int_t count const char ColorStruct_t color const char filename
R__EXTERN TSystem * gSystem
RooFit::OwningPtr< RooArgSet > getObservables(const RooArgSet &set, bool valueOnly=true) const
Given a set of possible observables, return the observables that this PDF depends on.
RooFit::OwningPtr< RooArgSet > getVariables(bool stripDisconnected=true) const
Return RooArgSet with all variables (tree leaf nodes of expresssion tree)
double getRealValue(const char *name, double defVal=0.0, bool verbose=false) const
Get value of a RooAbsReal stored in set with given name.
virtual bool add(const RooAbsArg &var, bool silent=false)
Add the specified argument to list.
RooAbsArg * first() const
void Print(Option_t *options=nullptr) const override
This method must be overridden when a class wants to print itself.
RooAbsData is the common abstract base class for binned and unbinned datasets.
virtual Int_t numEntries() const
Return number of entries in dataset, i.e., count unweighted entries.
RooFit::OwningPtr< RooDataSet > generate(const RooArgSet &whatVars, Int_t nEvents, const RooCmdArg &arg1, const RooCmdArg &arg2=RooCmdArg::none(), const RooCmdArg &arg3=RooCmdArg::none(), const RooCmdArg &arg4=RooCmdArg::none(), const RooCmdArg &arg5=RooCmdArg::none())
See RooAbsPdf::generate(const RooArgSet&,const RooCmdArg&,const RooCmdArg&,const RooCmdArg&,...
virtual RooFit::OwningPtr< RooAbsReal > createNLL(RooAbsData &data, const RooLinkedList &cmdList={})
Construct representation of -log(L) of PDF with given dataset.
bool canBeExtended() const
If true, PDF can provide extended likelihood term.
virtual double getMax(const char *name=nullptr) const
Get maximum of currently defined range.
virtual double getMin(const char *name=nullptr) const
Get minimum of currently defined range.
double getVal(const RooArgSet *normalisationSet=nullptr) const
Evaluate object.
RooArgSet is a container object that can hold multiple RooAbsArg objects.
TObject * clone(const char *newname) const override
RooArgSet * snapshot(bool deepCopy=true) const
Use RooAbsCollection::snapshot(), but return as RooArgSet.
RooDataSet is a container class to hold unbinned data.
const RooArgSet * get(Int_t index) const override
Return RooArgSet with coordinates of event 'index'.
RooRealVar represents a variable that can be changed from the outside.
void setVal(double value) override
Set value of variable to 'value'.
RooSimultaneous facilitates simultaneous fitting of multiple PDFs to subsets of a given dataset.
RooAbsPdf * getPdf(RooStringView catName) const
Return the p.d.f associated with the given index category name.
ConfidenceBelt is a concrete implementation of the ConfInterval interface.
double GetAcceptanceRegionMax(RooArgSet &, double cl=-1., double leftside=-1.)
The FeldmanCousins class (like the Feldman-Cousins technique) is essentially a specific configuration...
ModelConfig is a simple class that holds configuration information specifying how a model should be u...
const RooArgSet * GetGlobalObservables() const
get RooArgSet for global observables (return nullptr if not existing)
const RooArgSet * GetParametersOfInterest() const
get RooArgSet containing the parameter of interest (return nullptr if not existing)
const RooArgSet * GetNuisanceParameters() const
get RooArgSet containing the nuisance parameters (return nullptr if not existing)
const RooArgSet * GetObservables() const
get RooArgSet for observables (return nullptr if not existing)
RooAbsPdf * GetPdf() const
get model PDF (return nullptr if pdf has not been specified or does not exist)
PointSetInterval is a concrete implementation of the ConfInterval interface.
double UpperLimit(RooRealVar ¶m)
return upper limit on a given parameter
double LowerLimit(RooRealVar ¶m)
return lower limit on a given parameter
ProfileLikelihoodTestStat is an implementation of the TestStatistic interface that calculates the pro...
void SetOneSided(bool flag=true)
Holds configuration options for proof and proof-lite.
ToyMCSampler is an implementation of the TestStatSampler interface.
void SetProofConfig(ProofConfig *pc=nullptr)
calling with argument or nullptr deactivates proof
virtual TestStatistic * GetTestStatistic(unsigned int i) const
void SetGlobalObservables(const RooArgSet &o) override
specify the conditional observables
The RooWorkspace is a persistable container for RooFit projects.
A ROOT file is a suite of consecutive data records (TKey instances) with a well defined format.
static TFile * Open(const char *name, Option_t *option="", const char *ftitle="", Int_t compress=ROOT::RCompressionSetting::EDefaults::kUseCompiledDefault, Int_t netopt=0)
Create / open a file.
1-D histogram with a float per channel (see TH1 documentation)}
virtual Double_t GetBinCenter(Int_t bin) const
Return bin center for 1D histogram.
virtual Int_t GetNbinsX() const
virtual Int_t Fill(Double_t x)
Increment bin with abscissa X by 1.
virtual void SetContent(const Double_t *content)
Replace bin contents by the contents of array content.
void Draw(Option_t *option="") override
Draw this histogram with options.
virtual void SetMinimum(Double_t minimum=-1111)
virtual Double_t * GetIntegral()
Return a pointer to the array of bins integral.
TObject * Clone(const char *newname="") const override
Make a complete copy of the underlying object.
virtual void SetTitle(const char *title="")
Set the title of the TNamed.
const char * GetName() const override
Returns name of object.
virtual Bool_t AccessPathName(const char *path, EAccessMode mode=kFileExists)
Returns FALSE if one can access a file using the specified access mode.
RooCmdArg Extended(bool flag=true)
The namespace RooFit contains mostly switches that change the behaviour of functions of PDFs (or othe...
Namespace for the RooStats classes.
double SignificanceToPValue(double Z)
returns p-value corresponding to a 1-sided significance
- Authors
- Kyle Cranmer, Haichen Wang, Daniel Whiteson
Definition in file OneSidedFrequentistUpperLimitWithBands.C.


