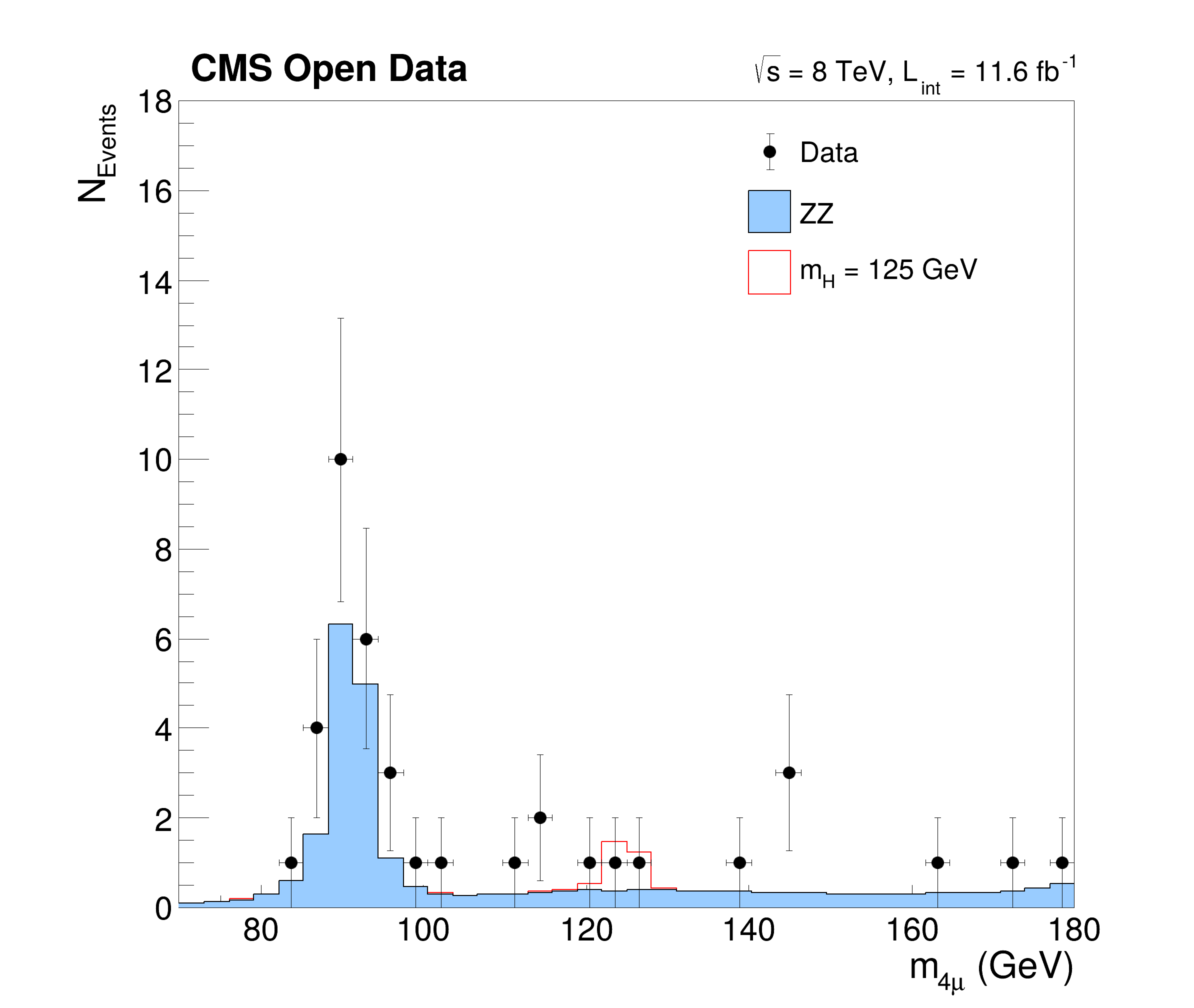
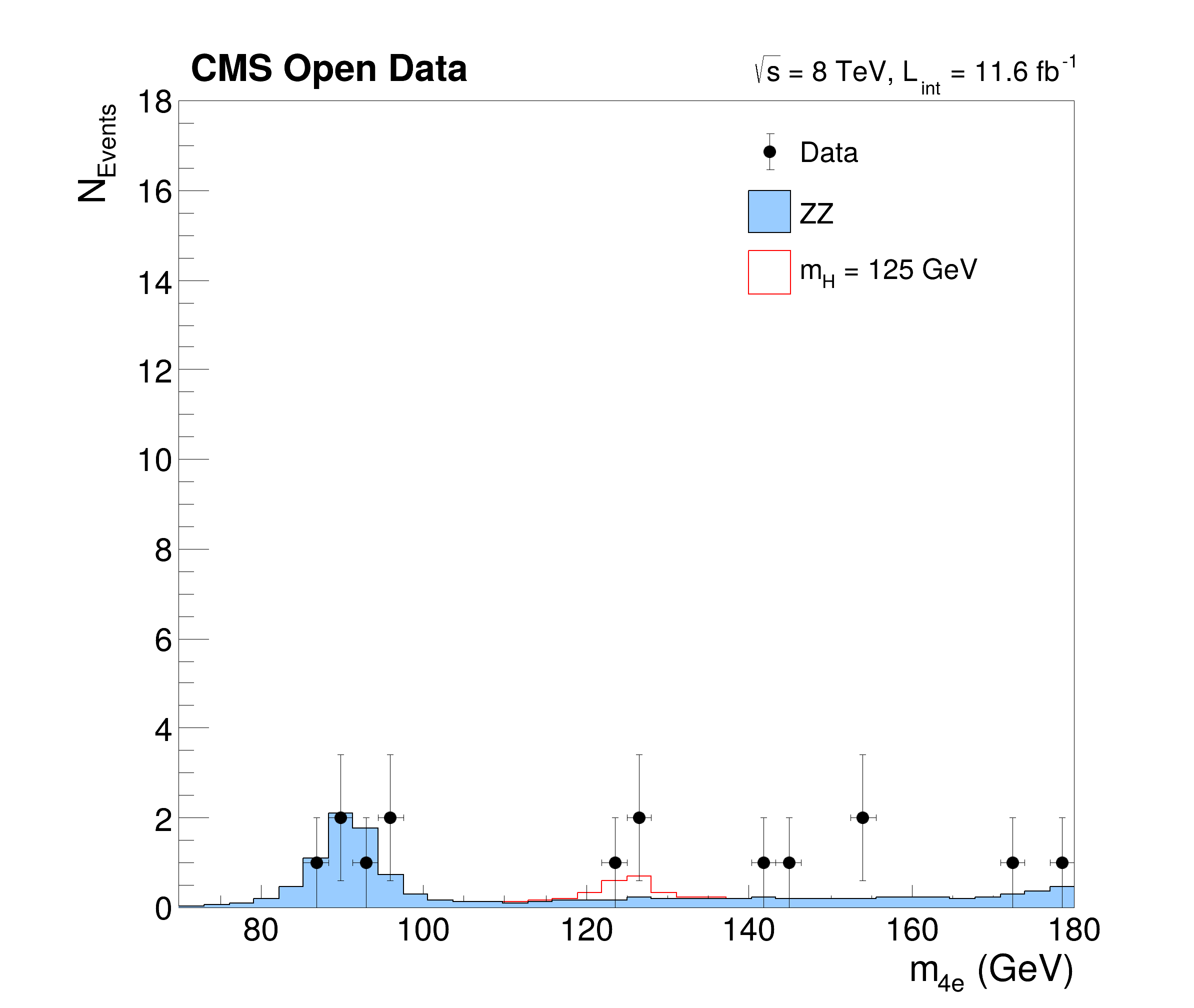
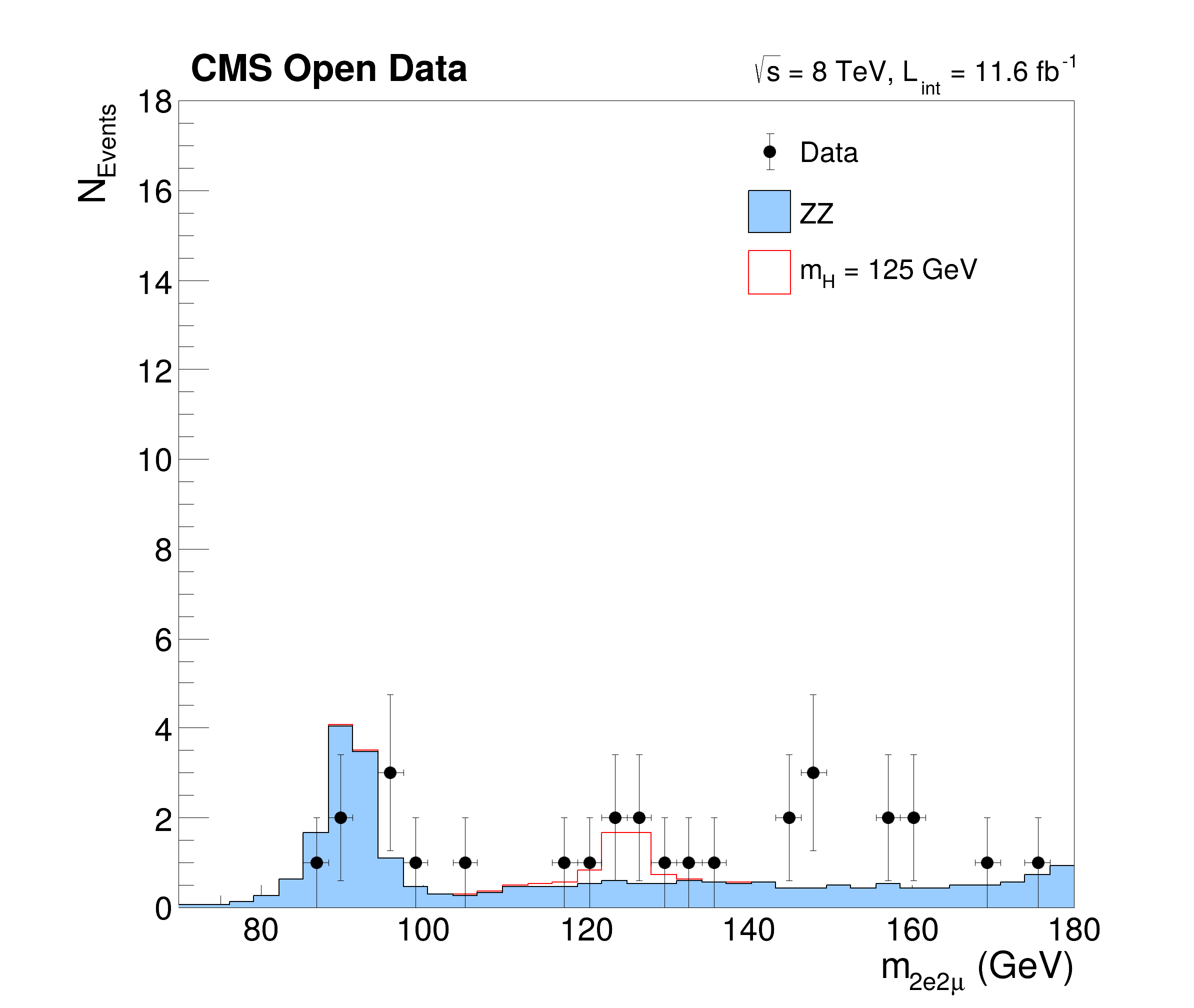
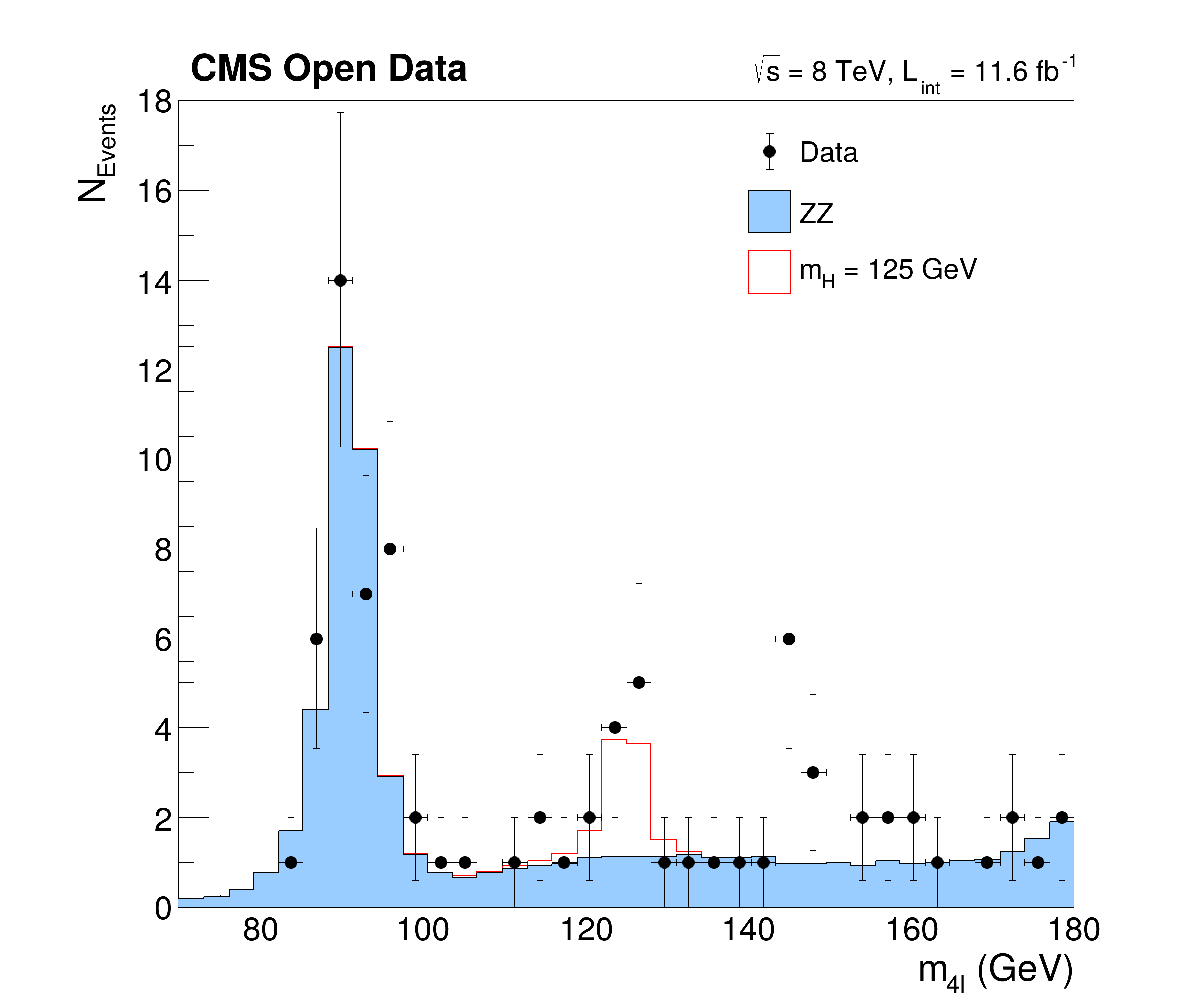
This tutorial is a simplified but yet complex example of an analysis reconstructing the Higgs boson decaying to two Z bosons from events with four leptons. The data and simulated events are taken from CERN OpenData representing a subset of the data recorded in 2012 with the CMS detector at the LHC. The tutorials follows the Higgs to four leptons analysis published on CERN Open Data portal (10.7483/OPENDATA.CMS.JKB8.RR42). The resulting plots show the invariant mass of the selected four lepton systems in different decay modes (four muons, four electrons and two of each kind) and in a combined plot indicating the decay of the Higgs boson with a mass of about 125 GeV.
The following steps are performed for each sample with data and simulated events in order to reconstruct the Higgs boson from the selected muons and electrons:
The tutorial has the fast mode enabled by default, which reads the data from already skimmed datasets with a total size of only 51MB. If the fast mode is disabled, the tutorial runs over the full dataset with a size of 12GB.
#include <string>
{
auto df_ge4m = df.Filter(
"nMuon>=4",
"At least four muons");
auto df_iso =
df_ge4m.Filter(
"All(abs(Muon_pfRelIso04_all)<0.40)",
"Require good isolation");
auto df_kin =
df_iso.Filter(
"All(Muon_pt>5) && All(abs(Muon_eta)<2.4)",
"Good muon kinematics");
auto df_ip3d =
df_kin.Define(
"Muon_ip3d",
"sqrt(Muon_dxy*Muon_dxy + Muon_dz*Muon_dz)");
auto df_sip3d =
df_ip3d.Define(
"Muon_sip3d",
"Muon_ip3d/sqrt(Muon_dxyErr*Muon_dxyErr + Muon_dzErr*Muon_dzErr)");
auto df_pv =
df_sip3d.Filter(
"All(Muon_sip3d<4) && All(abs(Muon_dxy)<0.5) && All(abs(Muon_dz)<1.0)",
"Track close to primary vertex with small uncertainty");
auto df_2p2n =
df_pv.Filter(
"nMuon==4 && Sum(Muon_charge==1)==2 && Sum(Muon_charge==-1)==2",
"Two positive and two negative muons");
}
{
auto df_ge4el = df.Filter(
"nElectron>=4",
"At least our electrons");
auto df_iso =
df_ge4el.Filter(
"All(abs(Electron_pfRelIso03_all)<0.40)",
"Require good isolation");
auto df_kin =
df_iso.Filter(
"All(Electron_pt>7) && All(abs(Electron_eta)<2.5)",
"Good Electron kinematics");
auto df_ip3d =
df_kin.Define(
"Electron_ip3d",
"sqrt(Electron_dxy*Electron_dxy + Electron_dz*Electron_dz)");
"Electron_ip3d/sqrt(Electron_dxyErr*Electron_dxyErr + Electron_dzErr*Electron_dzErr)");
auto df_pv =
df_sip3d.Filter(
"All(Electron_sip3d<4) && All(abs(Electron_dxy)<0.5) && "
"All(abs(Electron_dz)<1.0)",
"Track close to primary vertex with small uncertainty");
auto df_2p2n =
df_pv.Filter(
"nElectron==4 && Sum(Electron_charge==1)==2 && Sum(Electron_charge==-1)==2",
"Two positive and two negative electrons");
}
{
auto df_ge2el2mu = df.Filter(
"nElectron>=2 && nMuon>=2",
"At least two electrons and two muons");
auto df_eta =
df_ge2el2mu.Filter(
"All(abs(Electron_eta)<2.5) && All(abs(Muon_eta)<2.4)",
"Eta cuts");
return true;
}
return true;
}
return false;
};
return false;
}
return true;
};
auto df_dr =
df_pt.Filter(
dr_cuts, {
"Muon_eta",
"Muon_phi",
"Electron_eta",
"Electron_phi"},
"Dr cuts");
auto df_iso =
df_dr.Filter(
"All(abs(Electron_pfRelIso03_all)<0.40) && All(abs(Muon_pfRelIso04_all)<0.40)",
"Require good isolation");
auto df_el_ip3d =
df_iso.Define(
"Electron_ip3d_el",
"sqrt(Electron_dxy*Electron_dxy + Electron_dz*Electron_dz)");
"Electron_ip3d_el/sqrt(Electron_dxyErr*Electron_dxyErr + "
"Electron_dzErr*Electron_dzErr)");
auto df_el_track =
df_el_sip3d.Filter(
"All(Electron_sip3d_el<4) && All(abs(Electron_dxy)<0.5) && All(abs(Electron_dz)<1.0)",
"Electron track close to primary vertex with small uncertainty");
"Muon_ip3d_mu/sqrt(Muon_dxyErr*Muon_dxyErr + Muon_dzErr*Muon_dzErr)");
auto df_mu_track =
df_mu_sip3d.Filter(
"All(Muon_sip3d_mu<4) && All(abs(Muon_dxy)<0.5) && All(abs(Muon_dz)<1.0)",
"Muon track close to primary vertex with small uncertainty");
"Two opposite charged electron and muon pairs");
}
{
idx[0].reserve(2); idx[1].reserve(2);
for (
size_t i = 0; i <
idx_cmb[0].size(); i++) {
}
}
}
for (size_t i = 0; i < 4; i++) {
idx[1].emplace_back(i);
}
}
return idx;
}
{
for (size_t i = 0; i < 2; i++) {
const auto i1 = idx[i][0];
const auto i2 = idx[i][1];
}
} else {
}
}
{
const auto i1 = idx[0][0];
const auto i2 = idx[0][1];
const auto i3 = idx[1][0];
const auto i4 = idx[1][1];
}
{
auto df_z1_cut = df.Filter(
"Z_mass[0] > 40 && Z_mass[0] < 120",
"Mass of first Z candidate in [40, 120]");
auto df_z2_cut =
df_z1_cut.Filter(
"Z_mass[1] > 12 && Z_mass[1] < 120",
"Mass of second Z candidate in [12, 120]");
}
{
df_base.Define(
"Z_idx",
reco_zz_to_4l, {
"Muon_pt",
"Muon_eta",
"Muon_phi",
"Muon_mass",
"Muon_charge"});
for (size_t i = 0; i < 2; i++) {
const auto i1 = idx[i][0];
const auto i2 = idx[i][1];
return false;
}
}
return true;
};
df_z_idx.Filter(
filter_z_dr, {
"Z_idx",
"Muon_eta",
"Muon_phi"},
"Delta R separation of muons building Z system");
}
{
{"Electron_pt", "Electron_eta", "Electron_phi", "Electron_mass", "Electron_charge"});
for (size_t i = 0; i < 2; i++) {
const auto i1 = idx[i][0];
const auto i2 = idx[i][1];
return false;
}
}
return true;
};
"Delta R separation of Electrons building Z system");
{"Z_idx", "Electron_pt", "Electron_eta", "Electron_phi", "Electron_mass"});
{"Z_idx", "Electron_pt", "Electron_eta", "Electron_phi", "Electron_mass"});
}
{
} else {
}
}
{
}
{
"Muon_pt", "Muon_eta", "Muon_phi", "Muon_mass"});
{"Electron_pt", "Electron_eta", "Electron_phi", "Electron_mass", "Muon_pt", "Muon_eta", "Muon_phi", "Muon_mass"});
}
template <typename T>
{
auto h_cmb =
static_cast<TH1 *
>(sig->Clone());
h_cmb->GetXaxis()->SetTitleSize(0.04);
h_cmb->GetYaxis()->SetTitle(
"N_{Events}");
h_cmb->GetYaxis()->SetTitleSize(0.04);
h_bkg->SetFillStyle(1001);
h_bkg->Draw(
"HIST SAME");
cms_label.DrawLatexNDC(0.16, 0.92,
"#bf{CMS Open Data}");
header.
DrawLatexNDC(0.63, 0.92,
"#sqrt{s} = 8 TeV, L_{int} = 11.6 fb^{-1}");
}
{
std::string path = "root://eospublic.cern.ch//eos/root-eos/cms_opendata_2012_nanoaod/";
if (
run_fast) path =
"root://eospublic.cern.ch//eos/root-eos/cms_opendata_2012_nanoaod_skimmed/";
"Events", {path + "Run2012B_DoubleMuParked.root", path + "Run2012C_DoubleMuParked.root"});
"Events", {path + "Run2012B_DoubleElectron.root", path + "Run2012C_DoubleElectron.root"});
const auto nbins = 36;
.Histo1D({"h_sig_4mu", "", nbins, 70, 180}, "H_mass", "weight");
.Histo1D({"h_bkg_4mu", "", nbins, 70, 180}, "H_mass", "weight");
.Define("weight", [] { return 1.0; })
.Histo1D({"h_data_4mu", "", nbins, 70, 180}, "H_mass", "weight");
.Histo1D({"h_sig_4el", "", nbins, 70, 180}, "H_mass", "weight");
.Histo1D({"h_bkg_4el", "", nbins, 70, 180}, "H_mass", "weight");
.Histo1D({"h_data_4el", "", nbins, 70, 180}, "H_mass", "weight");
.Histo1D({"h_sig_2el2mu", "", nbins, 70, 180}, "H_mass", "weight");
.Histo1D({"h_bkg_2el2mu", "", nbins, 70, 180}, "H_mass", "weight");
.Histo1D({"h_data_2el2mu_doublemu", "", nbins, 70, 180}, "H_mass", "weight");
}
{
}
ROOT::Detail::TRangeCast< T, true > TRangeDynCast
TRangeDynCast is an adapter class that allows the typed iteration through a TCollection.
winID h TVirtualViewer3D TVirtualGLPainter char TVirtualGLPainter plot
Option_t Option_t TPoint TPoint const char GetTextMagnitude GetFillStyle GetLineColor GetLineWidth GetMarkerStyle GetTextAlign GetTextColor GetTextSize void data
Option_t Option_t TPoint TPoint const char GetTextMagnitude GetFillStyle GetLineColor GetLineWidth GetMarkerStyle GetTextAlign GetTextColor GetTextSize void char Point_t Rectangle_t WindowAttributes_t Float_t Float_t Float_t Int_t Int_t UInt_t UInt_t Rectangle_t Int_t Int_t Window_t TString Int_t GCValues_t GetPrimarySelectionOwner GetDisplay GetScreen GetColormap GetNativeEvent const char const char dpyName wid window const char font_name cursor keysym reg const char only_if_exist regb h Point_t winding char text const char depth char const char Int_t count const char ColorStruct_t color const char filename
R__EXTERN TStyle * gStyle
ROOT's RDataFrame offers a modern, high-level interface for analysis of data stored in TTree ,...
virtual void SetTextFont(Font_t tfont=62)
Set the text font.
virtual void SetTextSize(Float_t tsize=1)
Set the text size.
TH1 is the base class of all histogram classes in ROOT.
To draw Mathematical Formula.
TLatex * DrawLatexNDC(Double_t x, Double_t y, const char *text)
Draw this TLatex with new coordinates in NDC.
This class displays a legend box (TPaveText) containing several legend entries.
virtual void SaveAs(const char *filename="", Option_t *option="") const
Save this object in the file specified by filename.
RVec< T > Reverse(const RVec< T > &v)
Return copy of reversed vector.
RVec< RVec< std::size_t > > Combinations(const std::size_t size1, const std::size_t size2)
Return the indices that represent all combinations of the elements of two RVecs.
Vector1::Scalar DeltaR(const Vector1 &v1, const Vector2 &v2)
Find difference in pseudorapidity (Eta) and Phi between two generic vectors The only requirements on ...
RInterface<::ROOT::Detail::RDF::RNodeBase, void > RNode
unsigned int RunGraphs(std::vector< RResultHandle > handles)
Trigger the event loop of multiple RDataFrames concurrently.
void EnableImplicitMT(UInt_t numthreads=0)
Enable ROOT's implicit multi-threading for all objects and methods that provide an internal paralleli...
void Sort(Index n, const Element *a, Index *index, Bool_t down=kTRUE)
Sort the n elements of the array a of generic templated type Element.