| FourBinInstructional.C: This example is a generalization of the on/off problem. | Roostats tutorials | HybridOriginalDemo.C: Example on how to use the HybridCalculatorOriginal class |
HybridInstructional.C: example demostrating usage of HybridCalcultor
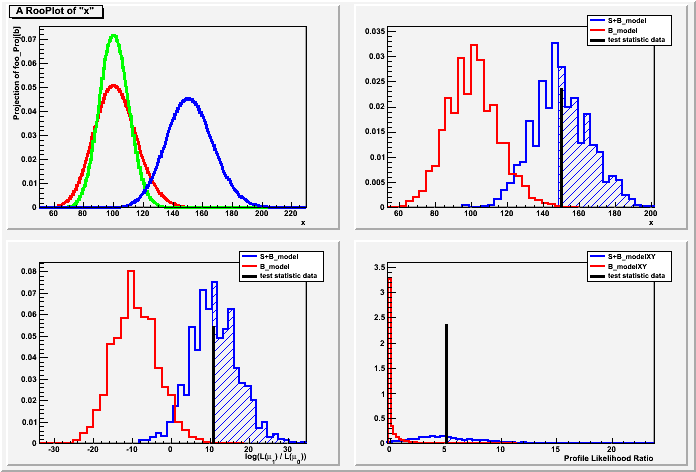
//+ example demostrating usage of HybridCalcultor /* HybridInstructional Authors: Kyle Cranmer, Wouter Verkerke, and Sven Kreiss date May 2010 Part 1-3 date Dec 2010 Part 4-6 A hypothesis testing example based on number counting with background uncertainty. NOTE: This example must be run with the ACLIC (the + option ) due to the new class that is defined. This example: - demonstrates the usage of the HybridCalcultor (Part 4-6) - demonstrates the numerical integration of RooFit (Part 2) - validates the RooStats against an example with a known analytic answer - demonstrates usage of different test statistics - explains subtle choices in the prior used for hybrid methods - demonstrates usage of different priors for the nuisance parameters - demonstrates usage of PROOF The basic setup here is that a main measurement has observed x events with an expectation of s+b. One can choose an ad hoc prior for the uncertainty on b, or try to base it on an auxiliary measurement. In this case, the auxiliary measurement (aka control measurement, sideband) is another counting experiment with measurement y and expectation tau*b. With an 'original prior' on b, called \eta(b) then one can obtain a posterior from the auxiliary measurement \pi(b) = \eta(b) * Pois(y|tau*b). This is a principled choice for a prior on b in the main measurement of x, which can then be treated in a hybrid Bayesian/Frequentist way. Additionally, one can try to treat the two measurements simultaneously, which is detailed in Part 6 of the tutorial. This tutorial is related to the FourBin.C tutorial in the modeling, but focuses on hypothesis testing instead of interval estimation. More background on this 'prototype problem' can be found in the following papers: Evaluation of three methods for calculating statistical significance when incorporating a systematic uncertainty into a test of the background-only hypothesis for a Poisson process Authors: Robert D. Cousins, James T. Linnemann, Jordan Tucker http://arxiv.org/abs/physics/0702156 NIM A 595 (2008) 480--501 Statistical Challenges for Searches for New Physics at the LHC Authors: Kyle Cranmer http://arxiv.org/abs/physics/0511028 Measures of Significance in HEP and Astrophysics Authors: J. T. Linnemann http://arxiv.org/abs/physics/0312059 */ #include "RooGlobalFunc.h" #include "RooRealVar.h" #include "RooProdPdf.h" #include "RooWorkspace.h" #include "RooDataSet.h" #include "TCanvas.h" #include "TStopwatch.h" #include "TH1.h" #include "RooPlot.h" #include "RooMsgService.h" #include "RooStats/NumberCountingUtils.h" #include "RooStats/HybridCalculator.h" #include "RooStats/ToyMCSampler.h" #include "RooStats/HypoTestPlot.h" #include "RooStats/ProfileLikelihoodTestStat.h" #include "RooStats/SimpleLikelihoodRatioTestStat.h" #include "RooStats/RatioOfProfiledLikelihoodsTestStat.h" #include "RooStats/MaxLikelihoodEstimateTestStat.h" using namespace RooFit; using namespace RooStats; ////////////////////////////////////////////////// // A New Test Statistic Class for this example. // It simply returns the sum of the values in a particular // column of a dataset. // You can ignore this class and focus on the macro below ////////////////////////////////////////////////// class BinCountTestStat : public TestStatistic { public: BinCountTestStat(void) : fColumnName("tmp") {} BinCountTestStat(string columnName) : fColumnName(columnName) {} virtual Double_t Evaluate(RooAbsData& data, RooArgSet& /*nullPOI*/) { // This is the main method in the interface Double_t value = 0.0; for(int i=0; i < data.numEntries(); i++) { value += data.get(i)->getRealValue(fColumnName.c_str()); } return value; } virtual const TString GetVarName() const { return fColumnName; } private: string fColumnName; protected: ClassDef(BinCountTestStat,1) }; ClassImp(BinCountTestStat) ////////////////////////////////////////////////// // The Actual Tutorial Macro ////////////////////////////////////////////////// void HybridInstructional() { // This tutorial has 6 parts // Table of Contents // Setup // 1. Make the model for the 'prototype problem' // Special cases // 2. Use RooFit's direct integration to get p-value & significance // 3. Use RooStats analytic solution for this problem // RooStats HybridCalculator -- can be generalized // 4. RooStats ToyMC version of 2. & 3. // 5. RooStats ToyMC with an equivalent test statistic // 6. RooStats ToyMC with simultaneous control & main measurement // It takes ~4 min without PROOF and ~2 min with PROOF on 4 cores. // Of course, everything looks nicer with more toys, which takes longer. #ifdef __CINT__ cout << "DO NOT RUN WITH CINT: we are using a custom test statistic "; cout << "which requires that this tutorial must be compiled "; cout << "with ACLIC" << endl; return; #endif TStopwatch t; t.Start(); TCanvas *c = new TCanvas; c->Divide(2,2); /////////////////////////////////////////////////////// // P A R T 1 : D I R E C T I N T E G R A T I O N ////////////////////////////////////////////////////// // Make model for prototype on/off problem // Pois(x | s+b) * Pois(y | tau b ) // for Z_Gamma, use uniform prior on b. RooWorkspace* w = new RooWorkspace("w"); w->factory("Poisson::px(x[150,0,500],sum::splusb(s[0,0,100],b[100,0,300]))"); w->factory("Poisson::py(y[100,0,500],prod::taub(tau[1.],b))"); w->factory("PROD::model(px,py)"); w->factory("Uniform::prior_b(b)"); // We will control the output level in a few places to avoid // verbose progress messages. We start by keeping track // of the current threshold on messages. RooFit::MsgLevel msglevel = RooMsgService::instance().globalKillBelow(); // Use PROOF-lite on multi-core machines ProofConfig* pc = NULL; // uncomment below if you want to use PROOF // pc = new ProofConfig(*w, 4, "workers=4", kFALSE); // machine with 4 cores // pc = new ProofConfig(*w, 2, "workers=2", kFALSE); // machine with 2 cores /////////////////////////////////////////////////////// // P A R T 2 : D I R E C T I N T E G R A T I O N ////////////////////////////////////////////////////// // This is not the 'RooStats' way, but in this case the distribution // of the test statistic is simply x and can be calculated directly // from the PDF using RooFit's built-in integration. // Note, this does not generalize to situations in which the test statistic // depends on many events (rows in a dataset). // construct the Bayesian-averaged model (eg. a projection pdf) // p'(x|s) = \int db p(x|s+b) * [ p(y|b) * prior(b) ] w->factory("PROJ::averagedModel(PROD::foo(px|b,py,prior_b),b)") ; RooMsgService::instance().setGlobalKillBelow(RooFit::ERROR); // lower message level // plot it, red is averaged model, green is b known exactly, blue is s+b av model RooPlot* frame = w->var("x")->frame(Range(50,230)) ; w->pdf("averagedModel")->plotOn(frame,LineColor(kRed)) ; w->pdf("px")->plotOn(frame,LineColor(kGreen)) ; w->var("s")->setVal(50.); w->pdf("averagedModel")->plotOn(frame,LineColor(kBlue)) ; c->cd(1); frame->Draw() ; w->var("s")->setVal(0.); // compare analytic calculation of Z_Bi // with the numerical RooFit implementation of Z_Gamma // for an example with x = 150, y = 100 // numeric RooFit Z_Gamma w->var("y")->setVal(100); w->var("x")->setVal(150); RooAbsReal* cdf = w->pdf("averagedModel")->createCdf(*w->var("x")); cdf->getVal(); // get ugly print messages out of the way cout << "-----------------------------------------"<<endl; cout << "Part 2" << endl; cout << "Hybrid p-value from direct integration = " << 1-cdf->getVal() << endl; cout << "Z_Gamma Significance = " << PValueToSignificance(1-cdf->getVal()) << endl; RooMsgService::instance().setGlobalKillBelow(msglevel); // set it back ///////////////////////////////////////////////// // P A R T 3 : A N A L Y T I C R E S U L T ///////////////////////////////////////////////// // In this special case, the integrals are known analytically // and they are implemented in RooStats::NumberCountingUtils // analytic Z_Bi double p_Bi = NumberCountingUtils::BinomialWithTauObsP(150, 100, 1); double Z_Bi = NumberCountingUtils::BinomialWithTauObsZ(150, 100, 1); cout << "-----------------------------------------"<<endl; cout << "Part 3" << endl; std::cout << "Z_Bi p-value (analytic): " << p_Bi << std::endl; std::cout << "Z_Bi significance (analytic): " << Z_Bi << std::endl; t.Stop(); t.Print(); t.Reset(); t.Start(); //////////////////////////////////////////////////////////////// // P A R T 4 : U S I N G H Y B R I D C A L C U L A T O R //////////////////////////////////////////////////////////////// // Now we demonstrate the RooStats HybridCalculator. // // Like all RooStats calculators it needs the data and a ModelConfig // for the relevant hypotheses. Since we are doing hypothesis testing // we need a ModelConfig for the null (background only) and the alternate // (signal+background) hypotheses. We also need to specify the PDF, // the parameters of interest, and the observables. Furthermore, since // the parameter of interest is floating, we need to specify which values // of the parameter corresponds to the null and alternate (eg. s=0 and s=50) // // define some sets of variables obs={x} and poi={s} // note here, x is the only observable in the main measurement // and y is treated as a separate measurement, which is used // to produce the prior that will be used in this calculation // to randomize the nuisance parameters. w->defineSet("obs","x"); w->defineSet("poi","s"); // create a toy dataset with the x=150 RooDataSet *data = new RooDataSet("d", "d", *w->set("obs")); data->add(*w->set("obs")); ////////////////////////////////////////////////////////// // Part 3a : Setup ModelConfigs // create the null (background-only) ModelConfig with s=0 ModelConfig b_model("B_model", w); b_model.SetPdf(*w->pdf("px")); b_model.SetObservables(*w->set("obs")); b_model.SetParametersOfInterest(*w->set("poi")); w->var("s")->setVal(0.0); // important! b_model.SetSnapshot(*w->set("poi")); // create the alternate (signal+background) ModelConfig with s=50 ModelConfig sb_model("S+B_model", w); sb_model.SetPdf(*w->pdf("px")); sb_model.SetObservables(*w->set("obs")); sb_model.SetParametersOfInterest(*w->set("poi")); w->var("s")->setVal(50.0); // important! sb_model.SetSnapshot(*w->set("poi")); ////////////////////////////////////////////////////////// // Part 3b : Choose Test Statistic // To make an equivalent calculation we need to use x as the test // statistic. This is not a built-in test statistic in RooStats // so we define it above. The new class inherits from the // RooStats::TestStatistic interface, and simply returns the value // of x in the dataset. BinCountTestStat binCount("x"); ////////////////////////////////////////////////////////// // Part 3c : Define Prior used to randomize nuisance parameters // // The prior used for the hybrid calculator is the posterior // from the auxiliary measurement y. The model for the aux. // measurement is Pois(y|tau*b), thus the likleihood function // is proportional to (has the form of) a Gamma distribution. // if the 'original prior' \eta(b) is uniform, then from // Bayes's theorem we have the posterior: // \pi(b) = Pois(y|tau*b) * \eta(b) // If \eta(b) is flat, then we arrive at a Gamma distribution. // Since RooFit will normalize the PDF we can actually supply // py=Pois(y,tau*b) that will be equivalent to multiplying by a uniform. // // Alternatively, we could explicitly use a gamma distribution: // w->factory("Gamma::gamma(b,sum::temp(y,1),1,0)"); // // or we can use some other ad hoc prior that do not naturally // follow from the known form of the auxiliary measurement. // The common choice is the equivlaent Gaussian: w->factory("Gaussian::gauss_prior(b,y, expr::sqrty('sqrt(y)',y))"); // this corresponds to the "Z_N" calculation. // // or one could use the analogous log-normal prior w->factory("Lognormal::lognorm_prior(b,y, expr::kappa('1+1./sqrt(y)',y))"); // // Ideally, the HybridCalculator would be able to inspect the full // model Pois(x | s+b) * Pois(y | tau b ) and be given the original // prior \eta(b) to form \pi(b) = Pois(y|tau*b) * \eta(b). // This is not yet implemented because in the general case // it is not easy to identify the terms in the PDF that correspond // to the auxiliary measurement. So for now, it must be set // explicitly with: // - ForcePriorNuisanceNull() // - ForcePriorNuisanceAlt() // the name "ForcePriorNuisance" was chosen because we anticipate // this to be auto-detected, but will leave the option open // to force to a different prior for the nuisance parameters. ////////////////////////////////////////////////////////// // Part 3d : Construct and configure the HybridCalculator HybridCalculator hc1(*data, sb_model, b_model); ToyMCSampler *toymcs1 = (ToyMCSampler*)hc1.GetTestStatSampler(); toymcs1->SetNEventsPerToy(1); // because the model is in number counting form toymcs1->SetTestStatistic(&binCount); // set the test statistic hc1.SetToys(20000,1000); hc1.ForcePriorNuisanceAlt(*w->pdf("py")); hc1.ForcePriorNuisanceNull(*w->pdf("py")); // if you wanted to use the ad hoc Gaussian prior instead // hc1.ForcePriorNuisanceAlt(*w->pdf("gauss_prior")); // hc1.ForcePriorNuisanceNull(*w->pdf("gauss_prior")); // if you wanted to use the ad hoc log-normal prior instead // hc1.ForcePriorNuisanceAlt(*w->pdf("lognorm_prior")); // hc1.ForcePriorNuisanceNull(*w->pdf("lognorm_prior")); // enable proof // NOTE: This test statistic is defined in this macro, and is not // working with PROOF currently. Luckily test stat is fast to evaluate. // if(pc) toymcs1->SetProofConfig(pc); // these lines save current msg level and then kill any messages below ERROR RooMsgService::instance().setGlobalKillBelow(RooFit::ERROR); // Get the result HypoTestResult *r1 = hc1.GetHypoTest(); RooMsgService::instance().setGlobalKillBelow(msglevel); // set it back cout << "-----------------------------------------"<<endl; cout << "Part 4" << endl; r1->Print(); t.Stop(); t.Print(); t.Reset(); t.Start(); c->cd(2); HypoTestPlot *p1 = new HypoTestPlot(*r1,30); // 30 bins, TS is discrete p1->Draw(); //////////////////////////////////////////////////////////////////////////// // P A R T 5 : U S I N G H Y B R I D C A L C U L A T O R W I T H // A N A L T E R N A T I V E T E S T S T A T I S T I C ///////////////////////////////////////////////////////////////////////////// // // A likelihood ratio test statistics should be 1-to-1 with the count x // when the value of b is fixed in the likelihood. This is implemented // by the SimpleLikelihoodRatioTestStat SimpleLikelihoodRatioTestStat slrts(*b_model.GetPdf(),*sb_model.GetPdf()); slrts.SetNullParameters(*b_model.GetSnapshot()); slrts.SetAltParameters(*sb_model.GetSnapshot()); // HYBRID CALCULATOR HybridCalculator hc2(*data, sb_model, b_model); ToyMCSampler *toymcs2 = (ToyMCSampler*)hc2.GetTestStatSampler(); toymcs2->SetNEventsPerToy(1); toymcs2->SetTestStatistic(&slrts); hc2.SetToys(20000,1000); hc2.ForcePriorNuisanceAlt(*w->pdf("py")); hc2.ForcePriorNuisanceNull(*w->pdf("py")); // if you wanted to use the ad hoc Gaussian prior instead // hc2.ForcePriorNuisanceAlt(*w->pdf("gauss_prior")); // hc2.ForcePriorNuisanceNull(*w->pdf("gauss_prior")); // if you wanted to use the ad hoc log-normal prior instead // hc2.ForcePriorNuisanceAlt(*w->pdf("lognorm_prior")); // hc2.ForcePriorNuisanceNull(*w->pdf("lognorm_prior")); // enable proof if(pc) toymcs2->SetProofConfig(pc); // these lines save current msg level and then kill any messages below ERROR RooMsgService::instance().setGlobalKillBelow(RooFit::ERROR); // Get the result HypoTestResult *r2 = hc2.GetHypoTest(); cout << "-----------------------------------------"<<endl; cout << "Part 5" << endl; r2->Print(); t.Stop(); t.Print(); t.Reset(); t.Start(); RooMsgService::instance().setGlobalKillBelow(msglevel); c->cd(3); HypoTestPlot *p2 = new HypoTestPlot(*r2,30); // 30 bins p2->Draw(); //////////////////////////////////////////////////////////////////////////// // P A R T 6 : U S I N G H Y B R I D C A L C U L A T O R W I T H // A N A L T E R N A T I V E T E S T S T A T I S T I C // A N D S I M U L T A N E O U S M O D E L ///////////////////////////////////////////////////////////////////////////// // // If one wants to use a test statistic in which the nuisance parameters // are profiled (in one way or another), then the PDF must constrain b. // Otherwise any observation x can always be explained with s=0 and b=x/tau. // // In this case, one is really thinking about the problem in a // different way. They are considering x,y simultaneously. // and the PDF should be Pois(x | s+b) * Pois(y | tau b ) // and the set 'obs' should be {x,y}. w->defineSet("obsXY","x,y"); // create a toy dataset with the x=150, y=100 w->var("x")->setVal(150.); w->var("y")->setVal(100.); RooDataSet *dataXY = new RooDataSet("dXY", "dXY", *w->set("obsXY")); dataXY->add(*w->set("obsXY")); // now we need new model configs, with PDF="model" ModelConfig b_modelXY("B_modelXY", w); b_modelXY.SetPdf(*w->pdf("model")); // IMPORTANT b_modelXY.SetObservables(*w->set("obsXY")); b_modelXY.SetParametersOfInterest(*w->set("poi")); w->var("s")->setVal(0.0); // IMPORTANT b_modelXY.SetSnapshot(*w->set("poi")); // create the alternate (signal+background) ModelConfig with s=50 ModelConfig sb_modelXY("S+B_modelXY", w); sb_modelXY.SetPdf(*w->pdf("model")); // IMPORTANT sb_modelXY.SetObservables(*w->set("obsXY")); sb_modelXY.SetParametersOfInterest(*w->set("poi")); w->var("s")->setVal(50.0); // IMPORTANT sb_modelXY.SetSnapshot(*w->set("poi")); // without this print, their can be a crash when using PROOF. Strange. // w->Print(); // Test statistics like the profile likelihood ratio // (or the ratio of profiled likelihoods (Tevatron) or the MLE for s) // will now work, since the nuisance parameter b is constrained by y. // ratio of alt and null likelihoods with background yield profiled. // // NOTE: These are slower because they have to run fits for each toy // Tevatron-style Ratio of profiled likelihoods // Q_Tev = -log L(s=0,\hat\hat{b})/L(s=50,\hat\hat{b}) RatioOfProfiledLikelihoodsTestStat ropl(*b_modelXY.GetPdf(), *sb_modelXY.GetPdf(), sb_modelXY.GetSnapshot()); ropl.SetSubtractMLE(false); // profile likelihood where alternate is best fit value of signal yield // \lambda(0) = -log L(s=0,\hat\hat{b})/L(\hat{s},\hat{b}) ProfileLikelihoodTestStat profll(*b_modelXY.GetPdf()); // just use the maximum likelihood estimate of signal yield // MLE = \hat{s} MaxLikelihoodEstimateTestStat mlets(*sb_modelXY.GetPdf(), *w->var("s")); // However, it is less clear how to justify the prior used in randomizing // the nuisance parameters (since that is a property of the ensemble, // and y is a property of each toy pseudo experiment. In that case, // one probably wants to consider a different y0 which will be held // constant and the prior \pi(b) = Pois(y0 | tau b) * \eta(b). w->factory("y0[100]"); w->factory("Gamma::gamma_y0(b,sum::temp0(y0,1),1,0)"); w->factory("Gaussian::gauss_prior_y0(b,y0, expr::sqrty0('sqrt(y0)',y0))"); // HYBRID CALCULATOR HybridCalculator hc3(*dataXY, sb_modelXY, b_modelXY); ToyMCSampler *toymcs3 = (ToyMCSampler*)hc3.GetTestStatSampler(); toymcs3->SetNEventsPerToy(1); toymcs3->SetTestStatistic(&slrts); hc3.SetToys(30000,1000); hc3.ForcePriorNuisanceAlt(*w->pdf("gamma_y0")); hc3.ForcePriorNuisanceNull(*w->pdf("gamma_y0")); // if you wanted to use the ad hoc Gaussian prior instead // hc3.ForcePriorNuisanceAlt(*w->pdf("gauss_prior_y0")); // hc3.ForcePriorNuisanceNull(*w->pdf("gauss_prior_y0")); // choose fit-based test statistic toymcs3->SetTestStatistic(&profll); //toymcs3->SetTestStatistic(&ropl); //toymcs3->SetTestStatistic(&mlets); // enable proof if(pc) toymcs3->SetProofConfig(pc); // these lines save current msg level and then kill any messages below ERROR RooMsgService::instance().setGlobalKillBelow(RooFit::ERROR); // Get the result HypoTestResult *r3 = hc3.GetHypoTest(); cout << "-----------------------------------------"<<endl; cout << "Part 6" << endl; r3->Print(); t.Stop(); t.Print(); t.Reset(); t.Start(); RooMsgService::instance().setGlobalKillBelow(msglevel); c->cd(4); c->GetPad(4)->SetLogy(); HypoTestPlot *p3 = new HypoTestPlot(*r3,50); // 50 bins p3->Draw(); c->SaveAs("zbi.pdf"); /////////////////////////////////////////////////////////// // OUTPUT W/O PROOF (2.66 GHz Intel Core i7) /////////////////////////////////////////////////////////// /* ----------------------------------------- Part 2 Hybrid p-value from direct integration = 0.00094165 Z_Gamma Significance = 3.10804 ----------------------------------------- Part 3 Z_Bi p-value (analytic): 0.00094165 Z_Bi significance (analytic): 3.10804 Real time 0:00:00, CP time 0.610 ----------------------------------------- Part 4 Results HybridCalculator_result: - Null p-value = 0.00115 +/- 0.000228984 - Significance = 3.04848 sigma - Number of S+B toys: 1000 - Number of B toys: 20000 - Test statistic evaluated on data: 150 - CL_b: 0.99885 +/- 0.000239654 - CL_s+b: 0.476 +/- 0.0157932 - CL_s: 0.476548 +/- 0.0158118 Real time 0:00:07, CP time 7.620 ----------------------------------------- Part 5 Results HybridCalculator_result: - Null p-value = 0.0009 +/- 0.000206057 - Significance = 3.12139 sigma - Number of S+B toys: 1000 - Number of B toys: 20000 - Test statistic evaluated on data: 10.8198 - CL_b: 0.9991 +/- 0.000212037 - CL_s+b: 0.465 +/- 0.0157726 - CL_s: 0.465419 +/- 0.0157871 Real time 0:00:34, CP time 34.360 ----------------------------------------- Part 6 Results HybridCalculator_result: - Null p-value = 0.000666667 +/- 0.000149021 - Significance = 3.20871 sigma - Number of S+B toys: 1000 - Number of B toys: 30000 - Test statistic evaluated on data: 5.03388 - CL_b: 0.999333 +/- 0.000149021 - CL_s+b: 0.511 +/- 0.0158076 - CL_s: 0.511341 +/- 0.0158183 Real time 0:05:06, CP time 306.330 */ /////////////////////////////////////////////////////////// // OUTPUT w/ PROOF (2.66 GHz Intel Core i7, 4 virtual cores) /////////////////////////////////////////////////////////// /* ----------------------------------------- Part 5 Results HybridCalculator_result: - Null p-value = 0.00075 +/- 0.000173124 - Significance = 3.17468 sigma - Number of S+B toys: 1000 - Number of B toys: 20000 - Test statistic evaluated on data: 10.8198 - CL_b: 0.99925 +/- 0.000193577 - CL_s+b: 0.454 +/- 0.0157443 - CL_s: 0.454341 +/- 0.0157564 Real time 0:00:16, CP time 0.990 ----------------------------------------- Part 6 Results HybridCalculator_result: - Null p-value = 0.0007 +/- 0.000152699 - Significance = 3.19465 sigma - Number of S+B toys: 1000 - Number of B toys: 30000 - Test statistic evaluated on data: 5.03388 - CL_b: 0.9993 +/- 0.000152699 - CL_s+b: 0.518 +/- 0.0158011 - CL_s: 0.518363 +/- 0.0158124 Real time 0:01:25, CP time 0.580 */ ////////////////////////////////////////// // Comparison /////////////////////////////////////////// // LEPStatToolsForLHC // https://plone4.fnal.gov:4430/P0/phystat/packages/0703002 // Uses Gaussian prior // CL_b = 6.218476e-04, Significance = 3.228665 sigma // ////////////////////////////////////////// // Comparison /////////////////////////////////////////// // Asymptotics // From the value of the profile likelihood ratio (5.0338) // The significance can be estimated using Wilks's theorem // significance = sqrt(2*profileLR) = 3.1729 sigma } HybridInstructional.C:1 HybridInstructional.C:2 HybridInstructional.C:3 HybridInstructional.C:4 HybridInstructional.C:5 HybridInstructional.C:6 HybridInstructional.C:7 HybridInstructional.C:8 HybridInstructional.C:9 HybridInstructional.C:10 HybridInstructional.C:11 HybridInstructional.C:12 HybridInstructional.C:13 HybridInstructional.C:14 HybridInstructional.C:15 HybridInstructional.C:16 HybridInstructional.C:17 HybridInstructional.C:18 HybridInstructional.C:19 HybridInstructional.C:20 HybridInstructional.C:21 HybridInstructional.C:22 HybridInstructional.C:23 HybridInstructional.C:24 HybridInstructional.C:25 HybridInstructional.C:26 HybridInstructional.C:27 HybridInstructional.C:28 HybridInstructional.C:29 HybridInstructional.C:30 HybridInstructional.C:31 HybridInstructional.C:32 HybridInstructional.C:33 HybridInstructional.C:34 HybridInstructional.C:35 HybridInstructional.C:36 HybridInstructional.C:37 HybridInstructional.C:38 HybridInstructional.C:39 HybridInstructional.C:40 HybridInstructional.C:41 HybridInstructional.C:42 HybridInstructional.C:43 HybridInstructional.C:44 HybridInstructional.C:45 HybridInstructional.C:46 HybridInstructional.C:47 HybridInstructional.C:48 HybridInstructional.C:49 HybridInstructional.C:50 HybridInstructional.C:51 HybridInstructional.C:52 HybridInstructional.C:53 HybridInstructional.C:54 HybridInstructional.C:55 HybridInstructional.C:56 HybridInstructional.C:57 HybridInstructional.C:58 HybridInstructional.C:59 HybridInstructional.C:60 HybridInstructional.C:61 HybridInstructional.C:62 HybridInstructional.C:63 HybridInstructional.C:64 HybridInstructional.C:65 HybridInstructional.C:66 HybridInstructional.C:67 HybridInstructional.C:68 HybridInstructional.C:69 HybridInstructional.C:70 HybridInstructional.C:71 HybridInstructional.C:72 HybridInstructional.C:73 HybridInstructional.C:74 HybridInstructional.C:75 HybridInstructional.C:76 HybridInstructional.C:77 HybridInstructional.C:78 HybridInstructional.C:79 HybridInstructional.C:80 HybridInstructional.C:81 HybridInstructional.C:82 HybridInstructional.C:83 HybridInstructional.C:84 HybridInstructional.C:85 HybridInstructional.C:86 HybridInstructional.C:87 HybridInstructional.C:88 HybridInstructional.C:89 HybridInstructional.C:90 HybridInstructional.C:91 HybridInstructional.C:92 HybridInstructional.C:93 HybridInstructional.C:94 HybridInstructional.C:95 HybridInstructional.C:96 HybridInstructional.C:97 HybridInstructional.C:98 HybridInstructional.C:99 HybridInstructional.C:100 HybridInstructional.C:101 HybridInstructional.C:102 HybridInstructional.C:103 HybridInstructional.C:104 HybridInstructional.C:105 HybridInstructional.C:106 HybridInstructional.C:107 HybridInstructional.C:108 HybridInstructional.C:109 HybridInstructional.C:110 HybridInstructional.C:111 HybridInstructional.C:112 HybridInstructional.C:113 HybridInstructional.C:114 HybridInstructional.C:115 HybridInstructional.C:116 HybridInstructional.C:117 HybridInstructional.C:118 HybridInstructional.C:119 HybridInstructional.C:120 HybridInstructional.C:121 HybridInstructional.C:122 HybridInstructional.C:123 HybridInstructional.C:124 HybridInstructional.C:125 HybridInstructional.C:126 HybridInstructional.C:127 HybridInstructional.C:128 HybridInstructional.C:129 HybridInstructional.C:130 HybridInstructional.C:131 HybridInstructional.C:132 HybridInstructional.C:133 HybridInstructional.C:134 HybridInstructional.C:135 HybridInstructional.C:136 HybridInstructional.C:137 HybridInstructional.C:138 HybridInstructional.C:139 HybridInstructional.C:140 HybridInstructional.C:141 HybridInstructional.C:142 HybridInstructional.C:143 HybridInstructional.C:144 HybridInstructional.C:145 HybridInstructional.C:146 HybridInstructional.C:147 HybridInstructional.C:148 HybridInstructional.C:149 HybridInstructional.C:150 HybridInstructional.C:151 HybridInstructional.C:152 HybridInstructional.C:153 HybridInstructional.C:154 HybridInstructional.C:155 HybridInstructional.C:156 HybridInstructional.C:157 HybridInstructional.C:158 HybridInstructional.C:159 HybridInstructional.C:160 HybridInstructional.C:161 HybridInstructional.C:162 HybridInstructional.C:163 HybridInstructional.C:164 HybridInstructional.C:165 HybridInstructional.C:166 HybridInstructional.C:167 HybridInstructional.C:168 HybridInstructional.C:169 HybridInstructional.C:170 HybridInstructional.C:171 HybridInstructional.C:172 HybridInstructional.C:173 HybridInstructional.C:174 HybridInstructional.C:175 HybridInstructional.C:176 HybridInstructional.C:177 HybridInstructional.C:178 HybridInstructional.C:179 HybridInstructional.C:180 HybridInstructional.C:181 HybridInstructional.C:182 HybridInstructional.C:183 HybridInstructional.C:184 HybridInstructional.C:185 HybridInstructional.C:186 HybridInstructional.C:187 HybridInstructional.C:188 HybridInstructional.C:189 HybridInstructional.C:190 HybridInstructional.C:191 HybridInstructional.C:192 HybridInstructional.C:193 HybridInstructional.C:194 HybridInstructional.C:195 HybridInstructional.C:196 HybridInstructional.C:197 HybridInstructional.C:198 HybridInstructional.C:199 HybridInstructional.C:200 HybridInstructional.C:201 HybridInstructional.C:202 HybridInstructional.C:203 HybridInstructional.C:204 HybridInstructional.C:205 HybridInstructional.C:206 HybridInstructional.C:207 HybridInstructional.C:208 HybridInstructional.C:209 HybridInstructional.C:210 HybridInstructional.C:211 HybridInstructional.C:212 HybridInstructional.C:213 HybridInstructional.C:214 HybridInstructional.C:215 HybridInstructional.C:216 HybridInstructional.C:217 HybridInstructional.C:218 HybridInstructional.C:219 HybridInstructional.C:220 HybridInstructional.C:221 HybridInstructional.C:222 HybridInstructional.C:223 HybridInstructional.C:224 HybridInstructional.C:225 HybridInstructional.C:226 HybridInstructional.C:227 HybridInstructional.C:228 HybridInstructional.C:229 HybridInstructional.C:230 HybridInstructional.C:231 HybridInstructional.C:232 HybridInstructional.C:233 HybridInstructional.C:234 HybridInstructional.C:235 HybridInstructional.C:236 HybridInstructional.C:237 HybridInstructional.C:238 HybridInstructional.C:239 HybridInstructional.C:240 HybridInstructional.C:241 HybridInstructional.C:242 HybridInstructional.C:243 HybridInstructional.C:244 HybridInstructional.C:245 HybridInstructional.C:246 HybridInstructional.C:247 HybridInstructional.C:248 HybridInstructional.C:249 HybridInstructional.C:250 HybridInstructional.C:251 HybridInstructional.C:252 HybridInstructional.C:253 HybridInstructional.C:254 HybridInstructional.C:255 HybridInstructional.C:256 HybridInstructional.C:257 HybridInstructional.C:258 HybridInstructional.C:259 HybridInstructional.C:260 HybridInstructional.C:261 HybridInstructional.C:262 HybridInstructional.C:263 HybridInstructional.C:264 HybridInstructional.C:265 HybridInstructional.C:266 HybridInstructional.C:267 HybridInstructional.C:268 HybridInstructional.C:269 HybridInstructional.C:270 HybridInstructional.C:271 HybridInstructional.C:272 HybridInstructional.C:273 HybridInstructional.C:274 HybridInstructional.C:275 HybridInstructional.C:276 HybridInstructional.C:277 HybridInstructional.C:278 HybridInstructional.C:279 HybridInstructional.C:280 HybridInstructional.C:281 HybridInstructional.C:282 HybridInstructional.C:283 HybridInstructional.C:284 HybridInstructional.C:285 HybridInstructional.C:286 HybridInstructional.C:287 HybridInstructional.C:288 HybridInstructional.C:289 HybridInstructional.C:290 HybridInstructional.C:291 HybridInstructional.C:292 HybridInstructional.C:293 HybridInstructional.C:294 HybridInstructional.C:295 HybridInstructional.C:296 HybridInstructional.C:297 HybridInstructional.C:298 HybridInstructional.C:299 HybridInstructional.C:300 HybridInstructional.C:301 HybridInstructional.C:302 HybridInstructional.C:303 HybridInstructional.C:304 HybridInstructional.C:305 HybridInstructional.C:306 HybridInstructional.C:307 HybridInstructional.C:308 HybridInstructional.C:309 HybridInstructional.C:310 HybridInstructional.C:311 HybridInstructional.C:312 HybridInstructional.C:313 HybridInstructional.C:314 HybridInstructional.C:315 HybridInstructional.C:316 HybridInstructional.C:317 HybridInstructional.C:318 HybridInstructional.C:319 HybridInstructional.C:320 HybridInstructional.C:321 HybridInstructional.C:322 HybridInstructional.C:323 HybridInstructional.C:324 HybridInstructional.C:325 HybridInstructional.C:326 HybridInstructional.C:327 HybridInstructional.C:328 HybridInstructional.C:329 HybridInstructional.C:330 HybridInstructional.C:331 HybridInstructional.C:332 HybridInstructional.C:333 HybridInstructional.C:334 HybridInstructional.C:335 HybridInstructional.C:336 HybridInstructional.C:337 HybridInstructional.C:338 HybridInstructional.C:339 HybridInstructional.C:340 HybridInstructional.C:341 HybridInstructional.C:342 HybridInstructional.C:343 HybridInstructional.C:344 HybridInstructional.C:345 HybridInstructional.C:346 HybridInstructional.C:347 HybridInstructional.C:348 HybridInstructional.C:349 HybridInstructional.C:350 HybridInstructional.C:351 HybridInstructional.C:352 HybridInstructional.C:353 HybridInstructional.C:354 HybridInstructional.C:355 HybridInstructional.C:356 HybridInstructional.C:357 HybridInstructional.C:358 HybridInstructional.C:359 HybridInstructional.C:360 HybridInstructional.C:361 HybridInstructional.C:362 HybridInstructional.C:363 HybridInstructional.C:364 HybridInstructional.C:365 HybridInstructional.C:366 HybridInstructional.C:367 HybridInstructional.C:368 HybridInstructional.C:369 HybridInstructional.C:370 HybridInstructional.C:371 HybridInstructional.C:372 HybridInstructional.C:373 HybridInstructional.C:374 HybridInstructional.C:375 HybridInstructional.C:376 HybridInstructional.C:377 HybridInstructional.C:378 HybridInstructional.C:379 HybridInstructional.C:380 HybridInstructional.C:381 HybridInstructional.C:382 HybridInstructional.C:383 HybridInstructional.C:384 HybridInstructional.C:385 HybridInstructional.C:386 HybridInstructional.C:387 HybridInstructional.C:388 HybridInstructional.C:389 HybridInstructional.C:390 HybridInstructional.C:391 HybridInstructional.C:392 HybridInstructional.C:393 HybridInstructional.C:394 HybridInstructional.C:395 HybridInstructional.C:396 HybridInstructional.C:397 HybridInstructional.C:398 HybridInstructional.C:399 HybridInstructional.C:400 HybridInstructional.C:401 HybridInstructional.C:402 HybridInstructional.C:403 HybridInstructional.C:404 HybridInstructional.C:405 HybridInstructional.C:406 HybridInstructional.C:407 HybridInstructional.C:408 HybridInstructional.C:409 HybridInstructional.C:410 HybridInstructional.C:411 HybridInstructional.C:412 HybridInstructional.C:413 HybridInstructional.C:414 HybridInstructional.C:415 HybridInstructional.C:416 HybridInstructional.C:417 HybridInstructional.C:418 HybridInstructional.C:419 HybridInstructional.C:420 HybridInstructional.C:421 HybridInstructional.C:422 HybridInstructional.C:423 HybridInstructional.C:424 HybridInstructional.C:425 HybridInstructional.C:426 HybridInstructional.C:427 HybridInstructional.C:428 HybridInstructional.C:429 HybridInstructional.C:430 HybridInstructional.C:431 HybridInstructional.C:432 HybridInstructional.C:433 HybridInstructional.C:434 HybridInstructional.C:435 HybridInstructional.C:436 HybridInstructional.C:437 HybridInstructional.C:438 HybridInstructional.C:439 HybridInstructional.C:440 HybridInstructional.C:441 HybridInstructional.C:442 HybridInstructional.C:443 HybridInstructional.C:444 HybridInstructional.C:445 HybridInstructional.C:446 HybridInstructional.C:447 HybridInstructional.C:448 HybridInstructional.C:449 HybridInstructional.C:450 HybridInstructional.C:451 HybridInstructional.C:452 HybridInstructional.C:453 HybridInstructional.C:454 HybridInstructional.C:455 HybridInstructional.C:456 HybridInstructional.C:457 HybridInstructional.C:458 HybridInstructional.C:459 HybridInstructional.C:460 HybridInstructional.C:461 HybridInstructional.C:462 HybridInstructional.C:463 HybridInstructional.C:464 HybridInstructional.C:465 HybridInstructional.C:466 HybridInstructional.C:467 HybridInstructional.C:468 HybridInstructional.C:469 HybridInstructional.C:470 HybridInstructional.C:471 HybridInstructional.C:472 HybridInstructional.C:473 HybridInstructional.C:474 HybridInstructional.C:475 HybridInstructional.C:476 HybridInstructional.C:477 HybridInstructional.C:478 HybridInstructional.C:479 HybridInstructional.C:480 HybridInstructional.C:481 HybridInstructional.C:482 HybridInstructional.C:483 HybridInstructional.C:484 HybridInstructional.C:485 HybridInstructional.C:486 HybridInstructional.C:487 HybridInstructional.C:488 HybridInstructional.C:489 HybridInstructional.C:490 HybridInstructional.C:491 HybridInstructional.C:492 HybridInstructional.C:493 HybridInstructional.C:494 HybridInstructional.C:495 HybridInstructional.C:496 HybridInstructional.C:497 HybridInstructional.C:498 HybridInstructional.C:499 HybridInstructional.C:500 HybridInstructional.C:501 HybridInstructional.C:502 HybridInstructional.C:503 HybridInstructional.C:504 HybridInstructional.C:505 HybridInstructional.C:506 HybridInstructional.C:507 HybridInstructional.C:508 HybridInstructional.C:509 HybridInstructional.C:510 HybridInstructional.C:511 HybridInstructional.C:512 HybridInstructional.C:513 HybridInstructional.C:514 HybridInstructional.C:515 HybridInstructional.C:516 HybridInstructional.C:517 HybridInstructional.C:518 HybridInstructional.C:519 HybridInstructional.C:520 HybridInstructional.C:521 HybridInstructional.C:522 HybridInstructional.C:523 HybridInstructional.C:524 HybridInstructional.C:525 HybridInstructional.C:526 HybridInstructional.C:527 HybridInstructional.C:528 HybridInstructional.C:529 HybridInstructional.C:530 HybridInstructional.C:531 HybridInstructional.C:532 HybridInstructional.C:533 HybridInstructional.C:534 HybridInstructional.C:535 HybridInstructional.C:536 HybridInstructional.C:537 HybridInstructional.C:538 HybridInstructional.C:539 HybridInstructional.C:540 HybridInstructional.C:541 HybridInstructional.C:542 HybridInstructional.C:543 HybridInstructional.C:544 HybridInstructional.C:545 HybridInstructional.C:546 HybridInstructional.C:547 HybridInstructional.C:548 HybridInstructional.C:549 HybridInstructional.C:550 HybridInstructional.C:551 HybridInstructional.C:552 HybridInstructional.C:553 HybridInstructional.C:554 HybridInstructional.C:555 HybridInstructional.C:556 HybridInstructional.C:557 HybridInstructional.C:558 HybridInstructional.C:559 HybridInstructional.C:560 HybridInstructional.C:561 HybridInstructional.C:562 HybridInstructional.C:563 HybridInstructional.C:564 HybridInstructional.C:565 HybridInstructional.C:566 HybridInstructional.C:567 HybridInstructional.C:568 HybridInstructional.C:569 HybridInstructional.C:570 HybridInstructional.C:571 HybridInstructional.C:572 HybridInstructional.C:573 HybridInstructional.C:574 HybridInstructional.C:575 HybridInstructional.C:576 HybridInstructional.C:577 HybridInstructional.C:578 HybridInstructional.C:579 HybridInstructional.C:580 HybridInstructional.C:581 HybridInstructional.C:582 HybridInstructional.C:583 HybridInstructional.C:584 HybridInstructional.C:585 HybridInstructional.C:586 HybridInstructional.C:587 HybridInstructional.C:588 HybridInstructional.C:589 HybridInstructional.C:590 HybridInstructional.C:591 HybridInstructional.C:592 HybridInstructional.C:593 HybridInstructional.C:594 HybridInstructional.C:595 HybridInstructional.C:596 HybridInstructional.C:597 HybridInstructional.C:598 HybridInstructional.C:599 HybridInstructional.C:600 HybridInstructional.C:601 HybridInstructional.C:602 HybridInstructional.C:603 HybridInstructional.C:604 HybridInstructional.C:605 HybridInstructional.C:606 HybridInstructional.C:607 HybridInstructional.C:608 HybridInstructional.C:609 HybridInstructional.C:610 HybridInstructional.C:611 HybridInstructional.C:612 HybridInstructional.C:613 HybridInstructional.C:614 HybridInstructional.C:615 HybridInstructional.C:616 HybridInstructional.C:617 |
|