[#2] PROGRESS:HistFactory -- Getting histogram ./data/example.root:/data
[#2] INFO:HistFactory -- Opened input file: ./data/example.root:
[#2] PROGRESS:HistFactory -- Getting histogram ./data/example.root:/signal
[#2] PROGRESS:HistFactory -- Getting histogram ./data/example.root:/background1
[#2] PROGRESS:HistFactory -- Getting histogram ./data/example.root:/background1_statUncert
[#2] PROGRESS:HistFactory -- Getting histogram ./data/example.root:/background2
Measurement Name: meas OutputFilePrefix: ./results/example_UsingPy POI: SigXsecOverSM Lumi: 1 LumiRelErr: 0.1 BinLow: 0 BinHigh: 1 ExportOnly: 1
Constant Params: Lumi alpha_syst1
Channels:
Channel Name: channel1 InputFile:
Data:
InputFile: ./data/example.root HistoName: data HistoPath: HistoAddress: 0x55de2651c7e0
statErrorConfig:
RelErrorThreshold: 0.05 ConstraintType: Poisson
Samples:
Name: signal Channel: channel1 NormalizeByTheory: True StatErrorActivate: False
InputFile: ./data/example.root HistName: signal HistoPath: HistoAddress: 0x55de26df4300
Name: background1 Channel: channel1 NormalizeByTheory: True StatErrorActivate: False
InputFile: ./data/example.root HistName: background1 HistoPath: HistoAddress: 0x55de264afca0
StatError Activate: 1 InputFile: ./data/example.root HistName: background1_statUncert HistoPath: HistoAddress: 0x55de206dba20
Name: background2 Channel: channel1 NormalizeByTheory: True StatErrorActivate: False
InputFile: ./data/example.root HistName: background2 HistoPath: HistoAddress: 0x55de206df530
StatError Activate: 1 InputFile: ./data/example.root HistName: HistoPath: HistoAddress: 0
End of Channel channel1
[#2] INFO:HistFactory -- End Measurement: meas
[#2] INFO:HistFactory -- Making Model and Measurements (Fast) for measurement: meas
[#2] INFO:HistFactory -- using lumi = 1 and lumiError = 0.1 including bins between 0 and 1
[#2] INFO:HistFactory -- fixing the following parameters:
Lumi
alpha_syst1
[#2] INFO:HistFactory -- Creating the HistoToWorkspaceFactoryFast factory
[#2] INFO:HistFactory -- Setting preprocess functions
[#2] PROGRESS:HistFactory -- Starting to process channel: channel1
[#2] PROGRESS:HistFactory --
-----------------------------------------
Starting to process 'channel1' channel with 1 observables
-----------------------------------------
[#2] INFO:HistFactory -- making normFactor: SigXsecOverSM
[#2] INFO:HistFactory -- processing hist signal
[#2] INFO:HistFactory -- signal_channel1 has no variation histograms
[#2] INFO:HistFactory -- processing hist background1
[#2] INFO:HistFactory -- background1_channel1 has no variation histograms
[#2] INFO:HistFactory -- Sample: background1 to be included in Stat Error for channel channel1
[#2] INFO:HistFactory -- Using external histogram for Stat Errors for Channel: channel1 Sample: background1 Error Histogram: background1_statUncert
[#2] INFO:HistFactory -- processing hist background2
[#2] INFO:HistFactory -- background2_channel1 has no variation histograms
[#2] INFO:HistFactory -- Sample: background2 to be included in Stat Error for channel channel1
[#2] INFO:HistFactory -- Making Statistical Uncertainty Hist for Channel: channel1 Sample: background2
[#2] INFO:HistFactory -- Making Total Uncertainty for bin 1 Error = 5 CentralVal = 100 RelativeError = 0.05
[#2] INFO:HistFactory -- Making Total Uncertainty for bin 2 Error = 10 CentralVal = 100 RelativeError = 0.1
[#2] INFO:HistFactory -- About to create Constraint Terms from: mc_stat_channel1 params: (gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
[#2] INFO:HistFactory -- Using Poisson StatErrors in channel: channel1
[#2] INFO:HistFactory -- Creating constraint for: gamma_stat_channel1_bin_0. Type of constraint: 1
[#2] INFO:HistFactory -- Creating constraint for: gamma_stat_channel1_bin_1. Type of constraint: 1
[#2] PROGRESS:HistFactory --
-----------------------------------------
import model into workspace
-----------------------------------------
[#1] INFO:NumericIntegration -- RooRealIntegral::init(channel1_model_Int[obs_x_channel1]) using numeric integrator RooBinIntegrator to calculate Int(obs_x_channel1)
RooDataSet::AsimovData[obs_x_channel1,weight:binWeightAsimov] = 2 entries (230 weighted)
RooWorkspace(channel1) channel1 workspace contents
variables
---------
(Lumi,SigXsecOverSM,alpha_syst1,alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1,nom_alpha_syst1,nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1,nominalLumi,obs_x_channel1)
p.d.f.s
-------
RooGaussian::alpha_syst1Constraint[ x=alpha_syst1 mean=nom_alpha_syst1 sigma=1 ] = 1
RooGaussian::alpha_syst2Constraint[ x=alpha_syst2 mean=nom_alpha_syst2 sigma=1 ] = 1
RooGaussian::alpha_syst3Constraint[ x=alpha_syst3 mean=nom_alpha_syst3 sigma=1 ] = 1
RooRealSumPdf::channel1_model[ signal_channel1_scaleFactors * signal_channel1_shapes + background1_channel1_scaleFactors * background1_channel1_shapes + background2_channel1_scaleFactors * background2_channel1_shapes ] = 220/230
RooPoisson::gamma_stat_channel1_bin_0_constraint[ x=nom_gamma_stat_channel1_bin_0 mean=gamma_stat_channel1_bin_0_poisMean ] = 0.019943
RooPoisson::gamma_stat_channel1_bin_1_constraint[ x=nom_gamma_stat_channel1_bin_1 mean=gamma_stat_channel1_bin_1_poisMean ] = 0.039861
RooGaussian::lumiConstraint[ x=Lumi mean=nominalLumi sigma=0.1 ] = 1
RooProdPdf::model_channel1[ lumiConstraint * alpha_syst1Constraint * alpha_syst2Constraint * alpha_syst3Constraint * gamma_stat_channel1_bin_0_constraint * gamma_stat_channel1_bin_1_constraint * channel1_model(obs_x_channel1) ] = 0.174888
functions
--------
RooHistFunc::background1_channel1_Hist_alphanominal[ depList=(obs_x_channel1) ] = 0
RooStats::HistFactory::FlexibleInterpVar::background1_channel1_epsilon[ paramList=(alpha_syst2) ] = 1
RooProduct::background1_channel1_scaleFactors[ background1_channel1_epsilon * Lumi ] = 1
RooProduct::background1_channel1_shapes[ background1_channel1_Hist_alphanominal * mc_stat_channel1 * channel1_model_binWidth ] = 0
RooHistFunc::background2_channel1_Hist_alphanominal[ depList=(obs_x_channel1) ] = 100
RooStats::HistFactory::FlexibleInterpVar::background2_channel1_epsilon[ paramList=(alpha_syst3) ] = 1
RooProduct::background2_channel1_scaleFactors[ background2_channel1_epsilon * Lumi ] = 1
RooProduct::background2_channel1_shapes[ background2_channel1_Hist_alphanominal * mc_stat_channel1 * channel1_model_binWidth ] = 200
RooBinWidthFunction::channel1_model_binWidth[ HistFuncForBinWidth=signal_channel1_Hist_alphanominal ] = 2
RooProduct::gamma_stat_channel1_bin_0_poisMean[ gamma_stat_channel1_bin_0 * gamma_stat_channel1_bin_0_tau ] = 400
RooProduct::gamma_stat_channel1_bin_1_poisMean[ gamma_stat_channel1_bin_1 * gamma_stat_channel1_bin_1_tau ] = 100
ParamHistFunc::mc_stat_channel1[ ] = 1
RooHistFunc::signal_channel1_Hist_alphanominal[ depList=(obs_x_channel1) ] = 10
RooStats::HistFactory::FlexibleInterpVar::signal_channel1_epsilon[ paramList=(alpha_syst1) ] = 1
RooProduct::signal_channel1_scaleFactors[ signal_channel1_epsilon * SigXsecOverSM * Lumi ] = 1
RooProduct::signal_channel1_shapes[ signal_channel1_Hist_alphanominal * channel1_model_binWidth ] = 20
datasets
--------
RooDataSet::asimovData(obs_x_channel1)
RooDataSet::obsData(obs_x_channel1)
embedded datasets (in pdfs and functions)
-----------------------------------------
RooDataHist::signal_channel1_Hist_alphanominalDHist(obs_x_channel1)
RooDataHist::background1_channel1_Hist_alphanominalDHist(obs_x_channel1)
RooDataHist::background2_channel1_Hist_alphanominalDHist(obs_x_channel1)
named sets
----------
ModelConfig_GlobalObservables:(nominalLumi,nom_alpha_syst1,nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
ModelConfig_Observables:(obs_x_channel1)
constraintTerms:(lumiConstraint,alpha_syst1Constraint,alpha_syst2Constraint,alpha_syst3Constraint,gamma_stat_channel1_bin_0_constraint,gamma_stat_channel1_bin_1_constraint)
globalObservables:(nominalLumi,nom_alpha_syst1,nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
likelihoodTerms:(channel1_model)
observables:(obs_x_channel1)
observablesSet:(obs_x_channel1)
generic objects
---------------
RooStats::ModelConfig::ModelConfig
[#2] INFO:HistFactory -- Setting Parameter(s) of Interest as: SigXsecOverSM
=== Using the following for ModelConfig ===
Observables: RooArgSet:: = (obs_x_channel1)
Parameters of Interest: RooArgSet:: = (SigXsecOverSM)
Nuisance Parameters: RooArgSet:: = (alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
Global Observables: RooArgSet:: = (nominalLumi,nom_alpha_syst1,nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
PDF: RooProdPdf::model_channel1[ lumiConstraint * alpha_syst1Constraint * alpha_syst2Constraint * alpha_syst3Constraint * gamma_stat_channel1_bin_0_constraint * gamma_stat_channel1_bin_1_constraint * channel1_model(obs_x_channel1) ] = 0.174888
[#2] INFO:HistFactory -- Opening File to hold channel: ./results/example_UsingPy_channel1_meas_model.root
[#2] INFO:HistFactory -- About to write channel measurement to file
[#2] PROGRESS:HistFactory -- Writing sample: signal
[#2] PROGRESS:HistFactory -- Writing sample: background1
[#2] PROGRESS:HistFactory -- Writing sample: background2
[#2] PROGRESS:HistFactory -- Saved all histograms
[#2] PROGRESS:HistFactory -- Saved Measurement
[#2] PROGRESS:HistFactory -- Successfully wrote channel to file
[#2] INFO:HistFactory -- full list of observables:
(obs_x_channel1)
[#2] PROGRESS:HistFactory --
-----------------------------------------
Entering combination
-----------------------------------------
RooWorkspace(combined) combined contents
variables
---------
(channelCat,nom_alpha_syst1,nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1,nominalLumi,obs_x_channel1)
datasets
--------
RooDataSet::obsData(obs_x_channel1,channelCat)
named sets
----------
ModelConfig_GlobalObservables:(nominalLumi,nom_alpha_syst1,nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
ModelConfig_Observables:(obs_x_channel1,channelCat)
globalObservables:(nominalLumi,nom_alpha_syst1,nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
observables:(obs_x_channel1,channelCat)
[#2] PROGRESS:HistFactory --
-----------------------------------------
Importing combined model
-----------------------------------------
[#2] INFO:HistFactory -- setting Lumi constant
[#2] INFO:HistFactory -- setting alpha_syst1 constant
[#2] PROGRESS:HistFactory --
-----------------------------------------
create toy data
-----------------------------------------
[#1] INFO:NumericIntegration -- RooRealIntegral::init(channel1_model_Int[obs_x_channel1]) using numeric integrator RooBinIntegrator to calculate Int(obs_x_channel1)
RooDataSet::AsimovData0[obs_x_channel1,channelCat,weight:binWeightAsimov] = 2 entries (230 weighted)
[#2] INFO:HistFactory -- Setting Parameter(s) of Interest as: SigXsecOverSM
=== Using the following for ModelConfig ===
Observables: RooArgSet:: = (obs_x_channel1,channelCat)
Parameters of Interest: RooArgSet:: = (SigXsecOverSM)
Nuisance Parameters: RooArgSet:: = (alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
Global Observables: RooArgSet:: = (nominalLumi,nom_alpha_syst1,nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
PDF: RooSimultaneous::simPdf[ indexCat=channelCat channel1=model_channel1 ] = 0.190787
[#2] PROGRESS:HistFactory -- Writing combined workspace to file: ./results/example_UsingPy_combined_meas_model.root
[#2] PROGRESS:HistFactory -- Writing combined measurement to file: ./results/example_UsingPy_combined_meas_model.root
[#2] PROGRESS:HistFactory -- Writing sample: signal
[#2] PROGRESS:HistFactory -- Writing sample: background1
[#2] PROGRESS:HistFactory -- Writing sample: background2
[#2] PROGRESS:HistFactory -- Saved all histograms
[#2] PROGRESS:HistFactory -- Saved Measurement
[#1] INFO:Minimization -- p.d.f. provides expected number of events, including extended term in likelihood.
[#1] INFO:Minimization -- Including the following constraint terms in minimization: (lumiConstraint,alpha_syst1Constraint,alpha_syst2Constraint,alpha_syst3Constraint,gamma_stat_channel1_bin_0_constraint,gamma_stat_channel1_bin_1_constraint)
[#1] INFO:Minimization -- The following global observables have been defined and their values are taken from the model: (nominalLumi,nom_alpha_syst1,nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
[#1] INFO:Fitting -- RooAbsPdf::fitTo(simPdf) fixing normalization set for coefficient determination to observables in data
[#1] INFO:Fitting -- using generic CPU library compiled with no vectorizations
[#1] INFO:Fitting -- Creation of NLL object took 1.44801 ms
[#1] INFO:Fitting -- RooAddition::defaultErrorLevel(nll_simPdf_obsData) Summation contains a RooNLLVar, using its error level
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: activating const optimization
[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: deactivating const optimization
[#1] INFO:Minimization -- p.d.f. provides expected number of events, including extended term in likelihood.
[#1] INFO:Minimization -- Including the following constraint terms in minimization: (lumiConstraint,alpha_syst1Constraint,alpha_syst2Constraint,alpha_syst3Constraint,gamma_stat_channel1_bin_0_constraint,gamma_stat_channel1_bin_1_constraint)
[#1] INFO:Minimization -- The global observables are not defined , normalize constraints with respect to the parameters (Lumi,SigXsecOverSM,alpha_syst1,alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
[#1] INFO:Fitting -- RooAbsPdf::fitTo(simPdf) fixing normalization set for coefficient determination to observables in data
[#1] INFO:Fitting -- Creation of NLL object took 755.442 μs
[#1] INFO:NumericIntegration -- RooRealIntegral::init(gamma_stat_channel1_bin_1_constraint_Int[gamma_stat_channel1_bin_1]) using numeric integrator RooIntegrator1D to calculate Int(gamma_stat_channel1_bin_1)
[#1] INFO:NumericIntegration -- RooRealIntegral::init(gamma_stat_channel1_bin_0_constraint_Int[gamma_stat_channel1_bin_0]) using numeric integrator RooIntegrator1D to calculate Int(gamma_stat_channel1_bin_0)
[#1] INFO:Minimization -- RooProfileLL::evaluate(RooEvaluatorWrapper_Profile[SigXsecOverSM]) Creating instance of MINUIT
[#1] INFO:Fitting -- RooAddition::defaultErrorLevel(nll_simPdf_obsData) Summation contains a RooNLLVar, using its error level
[#1] INFO:Minimization -- RooProfileLL::evaluate(RooEvaluatorWrapper_Profile[SigXsecOverSM]) determining minimum likelihood for current configurations w.r.t all observable
[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
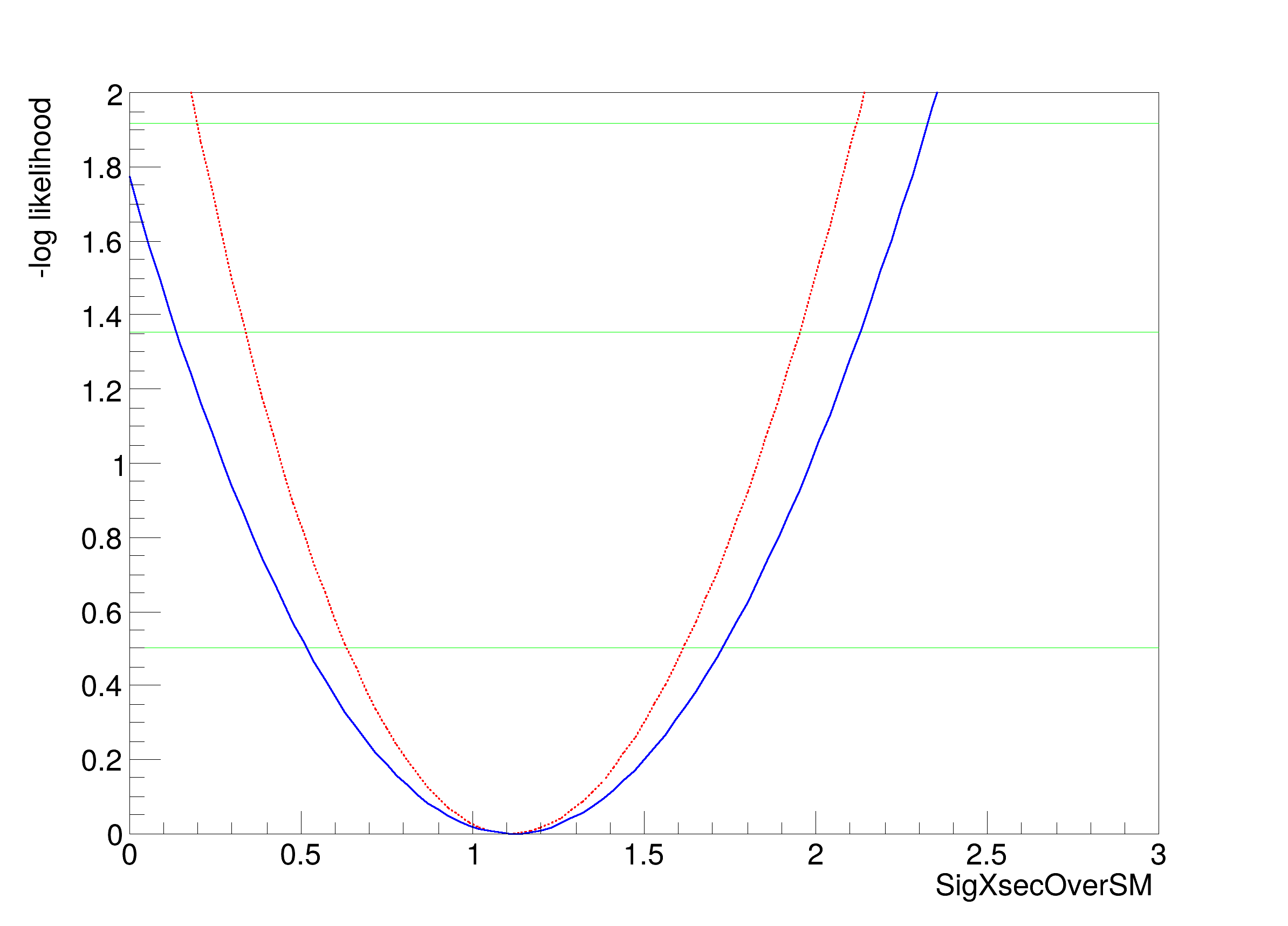
[#1] INFO:Minimization -- RooProfileLL::evaluate(RooEvaluatorWrapper_Profile[SigXsecOverSM]) minimum found at (SigXsecOverSM=1.11284)
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
.[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
RooFitResult: minimized FCN value: 15.1129, estimated distance to minimum: 0.000182975
covariance matrix quality: Full, accurate covariance matrix
Status : MINIMIZE=0 HESSE=0 MINOS=0
Constant Parameter Value
-------------------- ------------
Lumi 1.0000e+00
alpha_syst1 0.0000e+00
nom_alpha_syst1 0.0000e+00
nom_alpha_syst2 0.0000e+00
nom_alpha_syst3 0.0000e+00
nom_gamma_stat_channel1_bin_0 4.0000e+02
nom_gamma_stat_channel1_bin_1 1.0000e+02
nominalLumi 1.0000e+00
Floating Parameter InitialValue FinalValue (+HiError,-LoError) GblCorr.
-------------------- ------------ ---------------------------------- --------
SigXsecOverSM 1.0000e+00 1.1109e+00 (+6.16e-01,-5.93e-01) <none>
alpha_syst2 0.0000e+00 -1.0039e-02 (+9.90e-01,-9.88e-01) <none>
alpha_syst3 0.0000e+00 1.5339e-02 (+9.58e-01,-9.50e-01) <none>
gamma_stat_channel1_bin_0 1.0000e+00 9.9953e-01 (+5.03e-02,-4.86e-02) <none>
gamma_stat_channel1_bin_1 1.0000e+00 1.0050e+00 (+8.08e-02,-7.98e-02) <none>