'MULTIDIMENSIONAL MODELS' RooFit tutorial macro #303 Use of tailored p.d.f as conditional p.d.fs.s
pdf = gauss(x,f(y),sx | y ) with f(y) = a0 + a1*y
import ROOT
coord = {x, y}
if (abs(tmpy) < 10) and (abs(tmpx) < 10):
return d
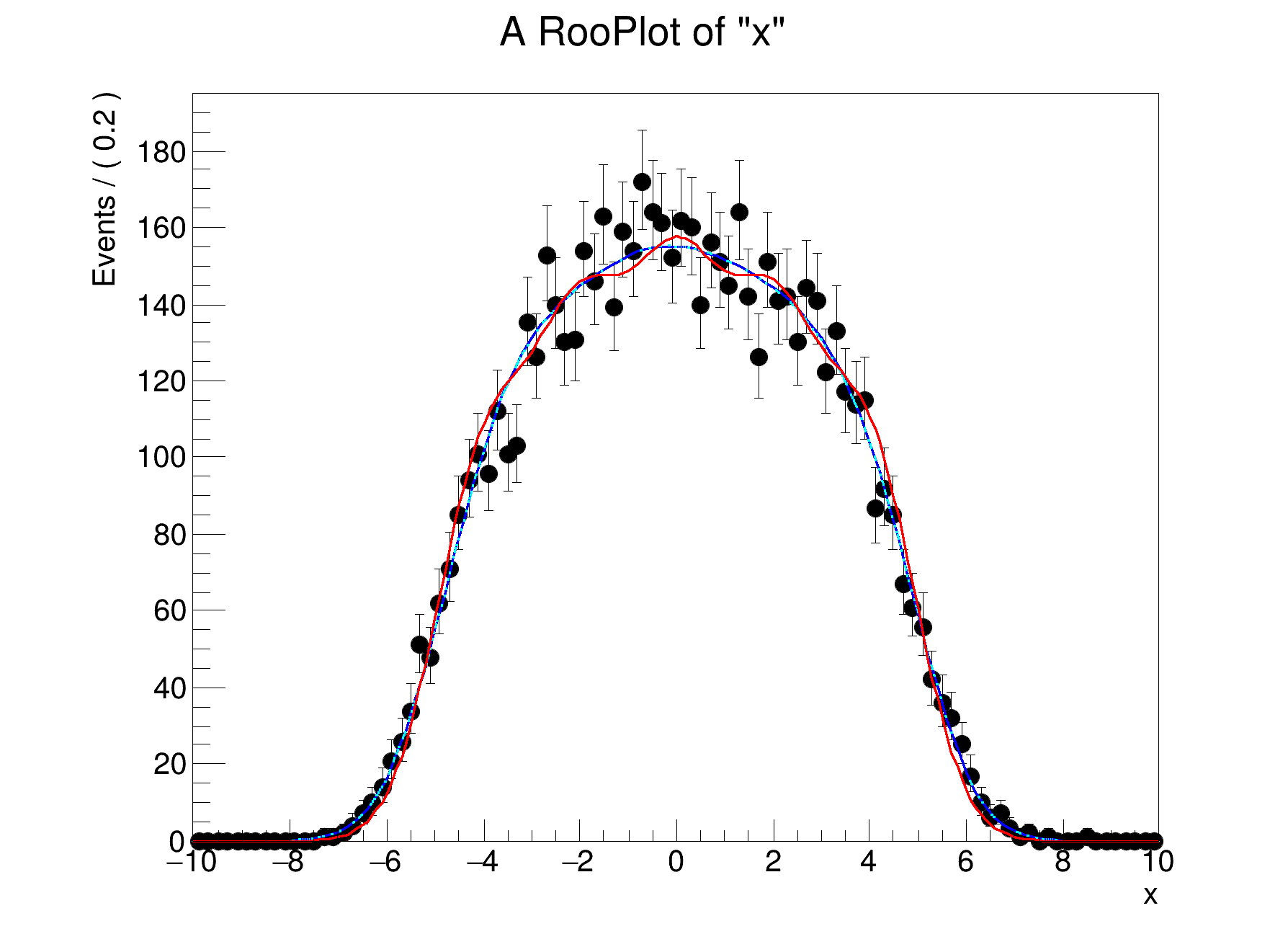
model.fitTo(expDataXY, ConditionalObservables={y}, PrintLevel=-1)
model.plotOn(xframe, ProjWData=binnedDataY, LineColor=
"c", LineStyle=
":")
c =
ROOT.TCanvas(
"rf303_conditional",
"rf303_conditional", 600, 460)
ROOT::Detail::TRangeCast< T, true > TRangeDynCast
TRangeDynCast is an adapter class that allows the typed iteration through a TCollection.
RooDataSet::modelData[x,y] = 6850 entries
[#1] INFO:Fitting -- RooAbsPdf::fitTo(model_over_model_Int[x]) fixing normalization set for coefficient determination to observables in data
[#1] INFO:Fitting -- using generic CPU library compiled with no vectorizations
[#1] INFO:Fitting -- Creation of NLL object took 837.503 μs
[#1] INFO:Fitting -- RooAddition::defaultErrorLevel(nll_model_over_model_Int[x]_d) Summation contains a RooNLLVar, using its error level
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: activating const optimization
[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: deactivating const optimization
[#1] INFO:Plotting -- RooAbsReal::plotOn(model) plot on x averages using data variables (y)
[#1] INFO:Plotting -- RooAbsReal::plotOn(model) plot on x averages using data variables (y)
[#1] INFO:Plotting -- RooAbsReal::plotOn(model) plot on x averages using data variables (y)
- Date
- February 2018
- Authors
- Clemens Lange, Wouter Verkerke (C version)
Definition in file rf303_conditional.py.