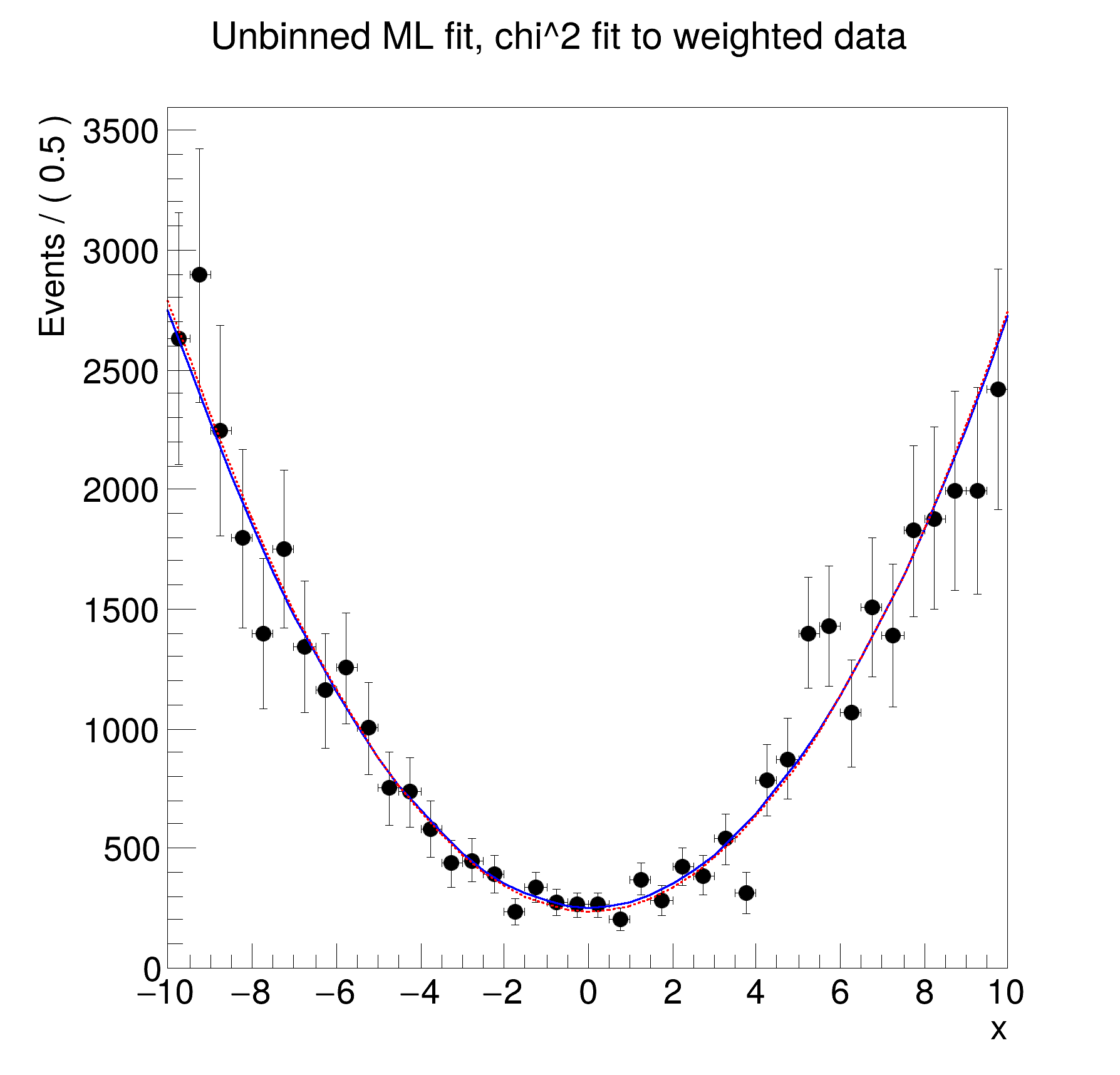
RooDataSet::pxData[x,w] = 1000 entries
RooDataSet::pxData[x,weight:w] = 1000 entries (43238.9 weighted)
[#1] INFO:Fitting -- RooAbsPdf::fitTo(p2_over_p2_Int[x]) fixing normalization set for coefficient determination to observables in data
[#1] INFO:Fitting -- using generic CPU library compiled with no vectorizations
[#1] INFO:Fitting -- Creation of NLL object took 945.972 μs
[#0] WARNING:InputArguments -- RooAbsPdf::fitTo(p2): WARNING: a likelihood fit is requested of what appears to be weighted data.
While the estimated values of the parameters will always be calculated taking the weights into account,
there are multiple ways to estimate the errors of the parameters. You are advised to make an
explicit choice for the error calculation:
- Either provide SumW2Error(true), to calculate a sum-of-weights-corrected HESSE error matrix
(error will be proportional to the number of events in MC).
- Or provide SumW2Error(false), to return errors from original HESSE error matrix
(which will be proportional to the sum of the weights, i.e., a dataset with <sum of weights> events).
- Or provide AsymptoticError(true), to use the asymptotically correct expression
(for details see https://arxiv.org/abs/1911.01303)."
[#1] INFO:Fitting -- RooAddition::defaultErrorLevel(nll_p2_over_p2_Int[x]_pxData) Summation contains a RooNLLVar, using its error level
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: activating const optimization
[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: deactivating const optimization
[#1] INFO:Fitting -- RooAbsPdf::fitTo(p2_over_p2_Int[x]) fixing normalization set for coefficient determination to observables in data
[#1] INFO:Fitting -- Creation of NLL object took 130.83 μs
[#1] INFO:Fitting -- RooAddition::defaultErrorLevel(nll_p2_over_p2_Int[x]_pxData) Summation contains a RooNLLVar, using its error level
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: activating const optimization
[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
[#1] INFO:Fitting -- RooAbsPdf::fitTo(p2) Calculating sum-of-weights-squared correction matrix for covariance matrix
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: deactivating const optimization
[#1] INFO:NumericIntegration -- RooRealIntegral::init(genPdf_Int[x]) using numeric integrator RooIntegrator1D to calculate Int(x)
[#1] INFO:NumericIntegration -- RooRealIntegral::init(genPdf_Int[x]) using numeric integrator RooIntegrator1D to calculate Int(x)
[#1] INFO:NumericIntegration -- RooRealIntegral::init(genPdf_Int[x]) using numeric integrator RooIntegrator1D to calculate Int(x)
[#1] INFO:NumericIntegration -- RooRealIntegral::init(genPdf_Int[x]) using numeric integrator RooIntegrator1D to calculate Int(x)
[#1] INFO:Fitting -- RooAbsPdf::fitTo(p2_over_p2_Int[x]) fixing normalization set for coefficient determination to observables in data
[#1] INFO:Fitting -- Creation of NLL object took 122.529 μs
[#1] INFO:Fitting -- RooAddition::defaultErrorLevel(nll_p2_over_p2_Int[x]_genPdfData) Summation contains a RooNLLVar, using its error level
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: activating const optimization
[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: deactivating const optimization
[#1] INFO:Fitting -- RooAbsPdf::fitTo(p2_over_p2_Int[x]) fixing normalization set for coefficient determination to observables in data
[#1] INFO:Fitting -- Creation of NLL object took 224.301 μs
[#1] INFO:Fitting -- RooAddition::defaultErrorLevel(nll_p2_over_p2_Int[x]_genPdfData) Summation contains a RooNLLVar, using its error level
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: activating const optimization
[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: deactivating const optimization
DataStore pxData_binned (Generated From px_binned)
Contains 40 entries
Observables:
1) x = 9.75 L(-10 - 10) B(40) "x"
Binned Dataset pxData_binned (Generated From px_binned)
Contains 40 bins with a total weight of 43238.9
Observables: 1) x = 9.75 L(-10 - 10) B(40) "x"
[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
Minuit2Minimizer: Minimize with max-calls 1000 convergence for edm < 1 strategy 1
Minuit2Minimizer : Valid minimum - status = 0
FVAL = 31.3747451817532266
Edm = 4.13511525432537654e-08
Nfcn = 29
a1 = -0.009989 +/- 0.0262975 (limited)
a2 = 0.106373 +/- 0.0101849 (limited)
==> ML Fit results on 1K unweighted events
RooFitResult: minimized FCN value: 2766.49, estimated distance to minimum: 0.000399952
covariance matrix quality: Full, accurate covariance matrix
Status : MINIMIZE=0 HESSE=0
Floating Parameter FinalValue +/- Error
-------------------- --------------------------
a1 8.9483e-03 +/- 2.70e-02
a2 1.0177e-01 +/- 1.69e-02
==> ML Fit results on 43K unweighted events
RooFitResult: minimized FCN value: 118892, estimated distance to minimum: 0.000206627
covariance matrix quality: Full, accurate covariance matrix
Status : MINIMIZE=0 HESSE=0
Floating Parameter FinalValue +/- Error
-------------------- --------------------------
a1 -1.2106e-03 +/- 4.02e-03
a2 9.7573e-02 +/- 2.37e-03
==> ML Fit results on 1K weighted events with a summed weight of 43K
RooFitResult: minimized FCN value: 119682, estimated distance to minimum: 1.25398e-05
covariance matrix quality: Full, accurate covariance matrix
Status : MINIMIZE=0 HESSE=0
Floating Parameter FinalValue +/- Error
-------------------- --------------------------
a1 -4.8713e-03 +/- 4.03e-03
a2 9.8645e-02 +/- 2.41e-03
==> Corrected ML Fit results on 1K weighted events with a summed weight of 43K
RooFitResult: minimized FCN value: 119682, estimated distance to minimum: 79498.5
covariance matrix quality: Full, accurate covariance matrix
Status : MINIMIZE=0 HESSE=0 HESSE=0
Floating Parameter FinalValue +/- Error
-------------------- --------------------------
a1 -4.8565e-03 +/- 3.00e-02
a2 9.8652e-02 +/- 2.99e-02
==> Chi2 Fit results on 1K weighted events with a summed weight of 43K
RooFitResult: minimized FCN value: 31.3747, estimated distance to minimum: 4.135e-08
covariance matrix quality: Full, accurate covariance matrix
Status : MIGRAD=0 HESSE=0
Floating Parameter FinalValue +/- Error
-------------------- --------------------------
a1 -9.9890e-03 +/- 2.63e-02
a2 1.0637e-01 +/- 1.02e-02