Processing /mnt/build/workspace/root-makedoc-v612/rootspi/rdoc/src/v6-12-00-patches/tutorials/roostats/StandardHypoTestDemo.C...
�[1mRooFit v3.60 -- Developed by Wouter Verkerke and David Kirkby�[0m
Copyright (C) 2000-2013 NIKHEF, University of California & Stanford University
All rights reserved, please read http://roofit.sourceforge.net/license.txt
RooWorkspace(combined) combined contents
variables
---------
(Lumi,SigXsecOverSM,alpha_syst1,alpha_syst2,alpha_syst3,binWidth_obs_x_channel1_0,binWidth_obs_x_channel1_1,binWidth_obs_x_channel1_2,channelCat,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1,nom_alpha_syst1,nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1,nominalLumi,obs_x_channel1,weightVar)
p.d.f.s
-------
RooGaussian::alpha_syst1Constraint[ x=alpha_syst1 mean=nom_alpha_syst1 sigma=1 ] = 1
RooGaussian::alpha_syst2Constraint[ x=alpha_syst2 mean=nom_alpha_syst2 sigma=1 ] = 1
RooGaussian::alpha_syst3Constraint[ x=alpha_syst3 mean=nom_alpha_syst3 sigma=1 ] = 1
RooRealSumPdf::channel1_model[ binWidth_obs_x_channel1_0 * L_x_signal_channel1_overallSyst_x_Exp + binWidth_obs_x_channel1_1 * L_x_background1_channel1_overallSyst_x_StatUncert + binWidth_obs_x_channel1_2 * L_x_background2_channel1_overallSyst_x_StatUncert ] = 220
RooPoisson::gamma_stat_channel1_bin_0_constraint[ x=nom_gamma_stat_channel1_bin_0 mean=gamma_stat_channel1_bin_0_poisMean ] = 0.019943
RooPoisson::gamma_stat_channel1_bin_1_constraint[ x=nom_gamma_stat_channel1_bin_1 mean=gamma_stat_channel1_bin_1_poisMean ] = 0.039861
RooGaussian::lumiConstraint[ x=Lumi mean=nominalLumi sigma=0.1 ] = 1
RooProdPdf::model_channel1[ lumiConstraint * alpha_syst1Constraint * alpha_syst2Constraint * alpha_syst3Constraint * gamma_stat_channel1_bin_0_constraint * gamma_stat_channel1_bin_1_constraint * channel1_model(obs_x_channel1) ] = 0.174888
RooSimultaneous::simPdf[ indexCat=channelCat channel1=model_channel1 ] = 0.174888
functions
--------
RooProduct::L_x_background1_channel1_overallSyst_x_StatUncert[ Lumi * background1_channel1_overallSyst_x_StatUncert ] = 0
RooProduct::L_x_background2_channel1_overallSyst_x_StatUncert[ Lumi * background2_channel1_overallSyst_x_StatUncert ] = 100
RooProduct::L_x_signal_channel1_overallSyst_x_Exp[ Lumi * signal_channel1_overallSyst_x_Exp ] = 10
RooStats::HistFactory::FlexibleInterpVar::background1_channel1_epsilon[ paramList=(alpha_syst2) ] = 1
RooHistFunc::background1_channel1_nominal[ depList=(obs_x_channel1) depList=(obs_x_channel1) ] = 0
RooProduct::background1_channel1_overallSyst_x_Exp[ background1_channel1_nominal * background1_channel1_epsilon ] = 0
RooProduct::background1_channel1_overallSyst_x_StatUncert[ mc_stat_channel1 * background1_channel1_overallSyst_x_Exp ] = 0
RooStats::HistFactory::FlexibleInterpVar::background2_channel1_epsilon[ paramList=(alpha_syst3) ] = 1
RooHistFunc::background2_channel1_nominal[ depList=(obs_x_channel1) depList=(obs_x_channel1) ] = 100
RooProduct::background2_channel1_overallSyst_x_Exp[ background2_channel1_nominal * background2_channel1_epsilon ] = 100
RooProduct::background2_channel1_overallSyst_x_StatUncert[ mc_stat_channel1 * background2_channel1_overallSyst_x_Exp ] = 100
RooProduct::gamma_stat_channel1_bin_0_poisMean[ gamma_stat_channel1_bin_0 * gamma_stat_channel1_bin_0_tau ] = 400
RooProduct::gamma_stat_channel1_bin_1_poisMean[ gamma_stat_channel1_bin_1 * gamma_stat_channel1_bin_1_tau ] = 100
ParamHistFunc::mc_stat_channel1[ ] = 1
RooStats::HistFactory::FlexibleInterpVar::signal_channel1_epsilon[ paramList=(alpha_syst1) ] = 1
RooHistFunc::signal_channel1_nominal[ depList=(obs_x_channel1) depList=(obs_x_channel1) ] = 10
RooProduct::signal_channel1_overallNorm_x_sigma_epsilon[ SigXsecOverSM * signal_channel1_epsilon ] = 1
RooProduct::signal_channel1_overallSyst_x_Exp[ signal_channel1_nominal * signal_channel1_overallNorm_x_sigma_epsilon ] = 10
datasets
--------
RooDataSet::asimovData(obs_x_channel1,weightVar,channelCat)
RooDataSet::obsData(channelCat,obs_x_channel1)
embedded datasets (in pdfs and functions)
-----------------------------------------
RooDataHist::signal_channel1nominalDHist(obs_x_channel1)
RooDataHist::background1_channel1nominalDHist(obs_x_channel1)
RooDataHist::background2_channel1nominalDHist(obs_x_channel1)
parameter snapshots
-------------------
NominalParamValues = (nom_alpha_syst2=0[C],nom_alpha_syst3=0[C],nom_gamma_stat_channel1_bin_0=400[C],nom_gamma_stat_channel1_bin_1=100[C],weightVar=0,obs_x_channel1=1.75,Lumi=1[C],nominalLumi=1[C],alpha_syst1=0[C],nom_alpha_syst1=0[C],alpha_syst2=0,alpha_syst3=0,gamma_stat_channel1_bin_0=1,gamma_stat_channel1_bin_1=1,SigXsecOverSM=1,binWidth_obs_x_channel1_0=2[C],binWidth_obs_x_channel1_1=2[C],binWidth_obs_x_channel1_2=2[C])
named sets
----------
ModelConfig_GlobalObservables:(nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
ModelConfig_NuisParams:(alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
ModelConfig_Observables:(obs_x_channel1,weightVar,channelCat)
ModelConfig_POI:(SigXsecOverSM)
globalObservables:(nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
observables:(obs_x_channel1,weightVar,channelCat)
generic objects
---------------
RooStats::ModelConfig::ModelConfig
=== Using the following for ModelConfigB_only ===
Observables: RooArgSet:: = (obs_x_channel1,weightVar,channelCat)
Parameters of Interest: RooArgSet:: = (SigXsecOverSM)
Nuisance Parameters: RooArgSet:: = (alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
Global Observables: RooArgSet:: = (nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
PDF: RooSimultaneous::simPdf[ indexCat=channelCat channel1=model_channel1 ] = 0.174888
Snapshot:
1) 0x47f75f0 RooRealVar:: SigXsecOverSM = 0 L(0 - 3) "SigXsecOverSM"
=== Using the following for ModelConfig ===
Observables: RooArgSet:: = (obs_x_channel1,weightVar,channelCat)
Parameters of Interest: RooArgSet:: = (SigXsecOverSM)
Nuisance Parameters: RooArgSet:: = (alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
Global Observables: RooArgSet:: = (nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
PDF: RooSimultaneous::simPdf[ indexCat=channelCat channel1=model_channel1 ] = 0.174888
Snapshot:
1) 0x47f75f0 RooRealVar:: SigXsecOverSM = 1 L(0 - 3) "SigXsecOverSM"
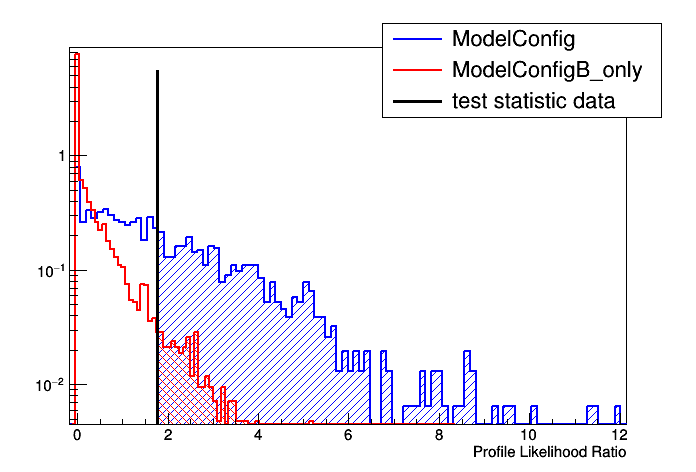
[#0] PROGRESS:Generation -- Test Statistic on data: 1.77404
[#1] INFO:InputArguments -- Profiling conditional MLEs for Null.
[#1] INFO:InputArguments -- Using a ToyMCSampler. Now configuring for Null.
[#0] PROGRESS:Generation -- generated toys: 500 / 5000
[#0] PROGRESS:Generation -- generated toys: 1000 / 5000
[#0] PROGRESS:Generation -- generated toys: 1500 / 5000
[#0] PROGRESS:Generation -- generated toys: 2000 / 5000
[#0] PROGRESS:Generation -- generated toys: 2500 / 5000
[#0] PROGRESS:Generation -- generated toys: 3000 / 5000
[#0] PROGRESS:Generation -- generated toys: 3500 / 5000
[#0] PROGRESS:Generation -- generated toys: 4000 / 5000
[#0] PROGRESS:Generation -- generated toys: 4500 / 5000
[#1] INFO:InputArguments -- Profiling conditional MLEs for Alt.
[#1] INFO:InputArguments -- Using a ToyMCSampler. Now configuring for Alt.
[#0] PROGRESS:Generation -- generated toys: 500 / 1250
[#0] PROGRESS:Generation -- generated toys: 1000 / 1250
Results HypoTestCalculator_result:
- Null p-value = 0.0304 +/- 0.002428
- Significance = 1.87495 +/- 0.0352942 sigma
- Number of Alt toys: 1250
- Number of Null toys: 5000
- Test statistic evaluated on data: 1.77404
- CL_b: 0.0304 +/- 0.002428
- CL_s+b: 0.4328 +/- 0.0140138
- CL_s: 14.2368 +/- 1.22696
Expected p -value and significance at -2 sigma = 0.839 significance -0.990356 sigma
Expected p -value and significance at -1 sigma = 0.2238 significance 0.759422 sigma
Expected p -value and significance at 0 sigma = 0.0434 significance 1.71252 sigma
Expected p -value and significance at 1 sigma = 0.0028 significance 2.77033 sigma
Expected p -value and significance at 2 sigma = 0.0002 significance 3.54008 sigma
#include <cassert>
struct HypoTestOptions {
bool noSystematics = false;
double nToysRatio = 4;
double poiValue = -1;
int printLevel=0;
bool generateBinned = false;
bool useProof = false;
bool enableDetailedOutput = false;
};
HypoTestOptions optHT;
void StandardHypoTestDemo(const char* infile = "",
const char* workspaceName = "combined",
const char* modelSBName = "ModelConfig",
const char* modelBName = "",
const char* dataName = "obsData",
int calcType = 0,
int testStatType = 3,
int ntoys = 5000,
bool useNC = false,
const char * nuisPriorName = 0)
{
bool noSystematics = optHT.noSystematics;
double nToysRatio = optHT.nToysRatio;
double poiValue = optHT.poiValue;
int printLevel = optHT.printLevel;
bool generateBinned = optHT.generateBinned;
bool useProof = optHT.useProof;
bool enableDetOutput = optHT.enableDetailedOutput;
SimpleLikelihoodRatioTestStat::SetAlwaysReuseNLL(true);
ProfileLikelihoodTestStat::SetAlwaysReuseNLL(true);
RatioOfProfiledLikelihoodsTestStat::SetAlwaysReuseNLL(true);
const char* filename = "";
if (!strcmp(infile,"")) {
filename = "results/example_combined_GaussExample_model.root";
if (!fileExist) {
#ifdef _WIN32
cout << "HistFactory file cannot be generated on Windows - exit" << endl;
return;
#endif
cout <<"will run standard hist2workspace example"<<endl;
gROOT->ProcessLine(
".! prepareHistFactory .");
gROOT->ProcessLine(
".! hist2workspace config/example.xml");
cout <<"\n\n---------------------"<<endl;
cout <<"Done creating example input"<<endl;
cout <<"---------------------\n\n"<<endl;
}
}
else
filename = infile;
if(!file ){
cout <<"StandardRooStatsDemoMacro: Input file " << filename << " is not found" << endl;
return;
}
if(!w){
cout <<"workspace not found" << endl;
return;
}
if(!data || !sbModel){
cout << "data or ModelConfig was not found" <<endl;
return;
}
if (noSystematics) {
if (nuisPar && nuisPar->
getSize() > 0) {
std::cout << "StandardHypoTestInvDemo" << " - Switch off all systematics by setting them constant to their initial values" << std::endl;
}
if (bModel) {
if (bnuisPar)
}
}
if (!bModel ) {
Info(
"StandardHypoTestInvDemo",
"The background model %s does not exist",modelBName);
Info(
"StandardHypoTestInvDemo",
"Copy it from ModelConfig %s and set POI to zero",modelSBName);
bModel->
SetName(TString(modelSBName)+TString(
"B_only"));
if (!var) return;
double oldval = var->
getVal();
}
Info(
"StandardHypoTestDemo",
"Model %s has no snapshot - make one using model poi",modelSBName);
if (!var) return;
double oldval = var->
getVal();
if (poiValue > 0) var->
setVal(poiValue);
if (poiValue > 0) var->
setVal(oldval);
}
if (enableDetOutput) {
ropl->EnableDetailedOutput();
}
AsymptoticCalculator::SetPrintLevel(printLevel);
else if (calcType == 1) hypoCalc=
new HybridCalculator(*data, *sbModel, *bModel);
if (calcType == 0) {
}
if (calcType == 1) {
}
if (calcType == 2 ) {
if (testStatType != 2 && testStatType != 3)
Warning(
"StandardHypoTestDemo",
"Only the PL test statistic can be used with AsymptoticCalculator - use by default a two-sided PL");
}
if (nuisPriorName) nuisPdf = w->
pdf(nuisPriorName);
if (!nuisPdf) {
Info(
"StandardHypoTestDemo",
"No nuisance pdf given for the HybridCalculator - try to deduce pdf from the model");
else
}
if (!nuisPdf ) {
Info(
"StandardHypoTestDemo",
"No nuisance pdf given - try to use %s that is defined as a prior pdf in the B model",nuisPdf->
GetName());
}
else {
Error(
"StandardHypoTestDemo",
"Cannot run Hybrid calculator because no prior on the nuisance parameter is specified or can be derived");
return;
}
}
assert(nuisPdf);
Info(
"StandardHypoTestDemo",
"Using as nuisance Pdf ... " );
Warning(
"StandardHypoTestDemo",
"Prior nuisance does not depend on nuisance parameters. They will be smeared in their full range");
}
delete np;
}
if (sampler && (calcType == 0 || calcType == 1) ) {
if (useNC)
Warning(
"StandardHypoTestDemo",
"Pdf is extended: but number counting flag is set: ignore it ");
}
else {
if (!useNC) {
Info(
"StandardHypoTestDemo",
"Pdf is not extended: number of events to generate taken from observed data set is %d",nEvents);
}
else {
Info(
"StandardHypoTestDemo",
"using a number counting pdf");
}
}
Info(
"StandardHypoTestDemo",
"Data set is weighted, nentries = %d and sum of weights = %8.1f but toy generation is unbinned - it would be faster to set generateBinned to true\n",data->
numEntries(), data->
sumEntries());
}
if (useProof) {
}
}
delete sampler;
delete slrts;
delete ropl;
delete profll;
if (calcType != 2) {
}
else {
std::cout << "Asymptotic results " << std::endl;
}
if (calcType != 2) {
htExp.Append(htr);
double p[5];
double q[5];
for (int i = 0; i < 5; ++i) {
double sig = -2 + i;
}
for (int i = 0; i < 5; ++i) {
htExp.SetTestStatisticData( q[i] );
double sig = -2 + i;
std::cout << " Expected p -value and significance at " << sig << " sigma = "
<< htExp.NullPValue() << " significance " << htExp.Significance() << " sigma " << std::endl;
}
}
else {
for (int i = 0; i < 5; ++i) {
double sig = -2 + i;
std::cout << " Expected p -value and significance at " << sig << " sigma = "
}
}
bool writeResult = (calcType != 2);
if (enableDetOutput) {
writeResult=true;
Info(
"StandardHypoTestDemo",
"Detailed output will be written in output result file");
}
if (htr != NULL && writeResult) {
const char * calcTypeName = (calcType == 0) ? "Freq" : (calcType == 1) ? "Hybr" : "Asym";
TString resultFileName =
TString::Format(
"%s_HypoTest_ts%d_",calcTypeName,testStatType);
TString name = infile;
name.Replace(0, name.Last('/')+1, "");
TFile * fileOut = new TFile(resultFileName,"RECREATE");
Info(
"StandardHypoTestDemo",
"HypoTestResult has been written in the file %s",resultFileName.Data());
fileOut->Close();
}
}



 Standard tutorial macro for hypothesis test (for computing the discovery significance) using all RooStats hypotheiss tests calculators and test statistics.
Standard tutorial macro for hypothesis test (for computing the discovery significance) using all RooStats hypotheiss tests calculators and test statistics.