A common method used in High Energy Physics to perform measurements is the maximum Likelihood method, exploiting discriminating variables to disentangle signal from background.
The crucial point for such an analysis to be reliable is to use an exhaustive list of sources of events combined with an accurate description of all the Probability Density Functions (PDF).
To assess the validity of the fit, a convincing quality check is to explore further the data sample by examining the distributions of control variables. A control variable can be obtained for instance by removing one of the discriminating variables before performing again the maximum Likelihood fit: this removed variable is a control variable. The expected distribution of this control variable, for signal, is to be compared to the one extracted, for signal, from the data sample. In order to be able to do so, one must be able to unfold from the distribution of the whole data sample.
The TSPlot method allows to reconstruct the distributions for the control variable, independently for each of the various sources of events, without making use of any a priori knowledge on this variable. The aim is thus to use the knowledge available for the discriminating variables to infer the behaviour of the individual sources of events with respect to the control variable.
TSPlot is optimal if the control variable is uncorrelated with the discriminating variables.
A detail description of the formalism itself, called \(\hbox{$_s$}{\cal P}lot\), is given in [1].
The \(\hbox{$_s$}{\cal P}lot\) technique is developed in the above context of a maximum Likelihood method making use of discriminating variables.
One considers a data sample in which are merged several species of events. These species represent various signal components and background components which all together account for the data sample. The different terms of the log-Likelihood are:
The extended log-Likelihood reads:
\[ {\cal L}=\sum_{e=1}^{N}\ln \Big\{ \sum_{i=1}^{{\rm N}_{\rm s}}N_i{\rm f}_i(y_e) \Big\} -\sum_{i=1}^{{\rm N}_{\rm s}}N_i \tag{1} \]
From this expression, after maximization of \({\cal L}\) with respect to the \(N_i\) parameters, a weight can be computed for every event and each species, in order to obtain later the true distribution \(\hbox{M}_i(x)\) of variable \(x\). If \({\rm n}\) is one of the \({\rm N}_{\rm s}\) species present in the data sample, the weight for this species is defined by:
\[ \fbox{$ {_s{\cal P}}_{\rm n}(y_e)={\sum_{j=1}^{{\rm N}_{\rm s}} \hbox{V}_{{\rm n}j}{\rm f}_j(y_e)\over\sum_{k=1}^{{\rm N}_{\rm s}}N_k{\rm f}_k(y_e) } $} , \tag{2} \]
where \(\hbox{V}_{{\rm n}j}\)
is the covariance matrix resulting from the Likelihood maximization. This matrix can be used directly from the fit, but this is numerically less accurate than the direct computation:
\[ \hbox{ V}^{-1}_{{\rm n}j}~=~ {\partial^2(-{\cal L})\over\partial N_{\rm n}\partial N_j}~=~ \sum_{e=1}^N {{\rm f}_{\rm n}(y_e){\rm f}_j(y_e)\over(\sum_{k=1}^{{\rm N}_{\rm s}}N_k{\rm f}_k(y_e))^2} . \tag{3} \]
The distribution of the control variable \(x\) obtained by histogramming the weighted events reproduces, on average, the true distribution \({\hbox{ {M}}}_{\rm n}(x)\)
The class TSPlot allows to reconstruct the true distribution \({\hbox{ {M}}}_{\rm n}(x)\)
of a control variable \(x\) for each of the \({\rm N}_{\rm s}\) species from the sole knowledge of the PDFs of the discriminating variables \({\rm f}_i(y)\). The plots obtained thanks to the TSPlot class are called \(\hbox {$_s$}{\cal P}lots\).
Beside reproducing the true distribution, \(\hbox {$_s$}{\cal P}lots\) bear remarkable properties:
\[ \sum_{e=1}^{N} {_s{\cal P}}_{\rm n}(y_e)~=~N_{\rm n} ~. \tag{4} \]
\[ \sum_{l=1}^{{\rm N}_{\rm s}} {_s{\cal P}}_l(y_e) ~=~1 ~. \tag{5} \]
That is to say that, summing up the \f${\rm N}_{\rm s}\f$ \f$\hbox {$_s$}{\cal P}lots\f$,
one recovers the data sample distribution in \f$x\f$, and summing up the number of events
entering in a \f$\hbox{$_s$}{\cal P}lot\f$ for a given species, one recovers the yield of the
species, as provided by the fit.
The property <a href="http://www.slac.stanford.edu/%7Epivk/sPlot/sPlot_ROOT/sPlot_ROOT.html#eq:NormalizationOK">4</a> is implemented in the TSPlot class as a check.
\[ \sigma[N_{\rm n}\ _s\tilde{\rm M}_{\rm n}(x) {\delta x}]~=~\sqrt{\sum_{e \subset {\delta x}} ({_s{\cal P}}_{\rm n})^2} ~. \tag{6} \]
reproduces the statistical uncertainty on the yield \f$N_{\rm n}\f$, as provided by the fit:
\f$\sigma[N_{\rm n}]\equiv\sqrt{\hbox{ V}_{{\rm n}{\rm n}}}\f$ .
Because of that and since the determination of the yields is optimal
when obtained using a Likelihood fit, one can conclude that the \f$\hbox{$_s$}{\cal P}lot\f$
technique is itself an optimal method to reconstruct distributions of control variables.
The \(\hbox {$_s$}{\cal P}lots\) reproduce the true distributions of the species in the control variable \(x\), within the above defined statistical uncertainties.
To illustrate the technique, one considers an example derived from the analysis where \(\hbox {$_s$}{\cal P}lots\) have been first used (charmless B decays). One is dealing with a data sample in which two species are present: the first is termed signal and the second background. A maximum Likelihood fit is performed to obtain the two yields \(N_1\) and \(N_2\) . The fit relies on two discriminating variables collectively denoted \(y\) which are chosen within three possible variables denoted \({m_{\rm ES}}\) , \(\Delta E\) and \({\cal F}\). The variable which is not incorporated in \(y\) is used as the control variable \(x\) . The six distributions of the three variables are assumed to be the ones depicted in Fig. 1.

Distributions of the three discriminating variables available to perform the Likelihood fit: \({m_{\rm ES}}\) , \(\Delta E\) , \({\cal F}\) . Among the three variables, two are used to perform the fit while one is kept out of the fit to serve the purpose of a control variable. The three distributions on the top (resp. bottom) of the figure correspond to the signal (resp. background). The unit of the vertical axis is chosen such that it indicates the number of entries per bin, if one slices the histograms in 25 bins.
A data sample being built through a Monte Carlo simulation based on the distributions shown in Fig. 1, one obtains the three distributions of Fig. 2. Whereas the distribution of \(\Delta E\) clearly indicates the presence of the signal, the distribution of \({m_{\rm ES}}\) and \({\cal F}\) are less obviously populated by signal.

Distributions of the three discriminating variables for signal plus background. The three distributions are the ones obtained from a data sample obtained through a Monte Carlo simulation based on the distributions shown in Fig. 1. The data sample consists of 500 signal events and 5000 background events.
Choosing \(\Delta E\) and \({\cal F}\) as discriminating variables to determine \(N_1\) and \(N_2\) through a maximum Likelihood fit, one builds, for the control variable \({m_{\rm ES}}\) which is unknown to the fit, the two \(\hbox {$_s$}{\cal P}lots\) for signal and background shown in Fig. 3. One observes that the \(\hbox{$_s$}{\cal P}lot\) for signal reproduces correctly the PDF even where the latter vanishes, although the error bars remain sizeable. This results from the almost complete cancellation between positive and negative weights: the sum of weights is close to zero while the sum of weights squared is not. The occurence of negative weights occurs through the appearance of the covariance matrix, and its negative components, in the definition of Eq. (2).
A word of caution is in order with respect to the error bars. Whereas their sum in quadrature is identical to the statistical uncertainties of the yields determined by the fit, and if, in addition, they are asymptotically correct, the error bars should be handled with care for low statistics and/or for too fine binning. This is because the error bars do not incorporate two known properties of the PDFs: PDFs are positive definite and can be non-zero in a given x-bin, even if in the particular data sample at hand, no event is observed in this bin. The latter limitation is not specific to \(\hbox {$_s$}{\cal P}lots\) , rather it is always present when one is willing to infer the PDF at the origin of an histogram, when, for some bins, the number of entries does not guaranty the applicability of the Gaussian regime. In such situations, a satisfactory practice is to attach allowed ranges to the histogram to indicate the upper and lower limits of the PDF value which are consistent with the actual observation, at a given confidence level.
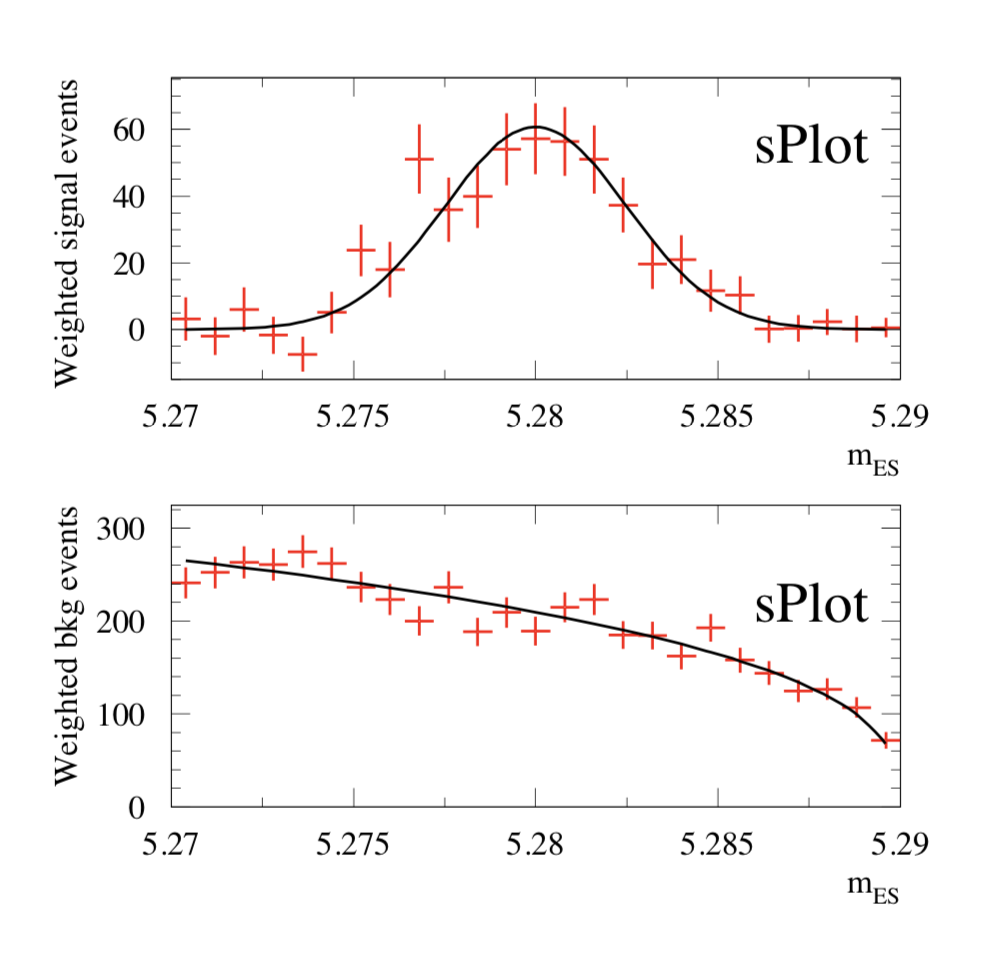
The \(\hbox {$_s$}{\cal P}lots\) (signal on top, background on bottom) obtained for \({m_{\rm ES}}\) are represented as dots with error bars. They are obtained from a fit using only information from \(\Delta E\) and \({\cal F}\)
Choosing \({m_{\rm ES}}\) and \(\Delta E\) as discriminating variables to determine \(N_1\) and \(N_2\) through a maximum Likelihood fit, one builds, for the control variable \({\cal F}\) which is unknown to the fit, the two \(\hbox {$_s$}{\cal P}lots\) for signal and background shown in Fig. 4. In the \(\hbox{$_s$}{\cal P}lot\) for signal one observes that error bars are the largest in the \(x\) regions where the background is the largest.

The \(\hbox {$_s$}{\cal P}lots\) (signal on top, background on bottom) obtained for \({\cal F}\) are represented as dots with error bars. They are obtained from a fit using only information from \({m_{\rm ES}}\) and \(\Delta E\)
The results above can be obtained by running the tutorial TestSPlot.C
Public Member Functions | |
| TSPlot () | |
| default constructor (used by I/O only) | |
| TSPlot (Int_t nx, Int_t ny, Int_t ne, Int_t ns, TTree *tree) | |
| Normal TSPlot constructor. | |
| virtual | ~TSPlot () |
| Destructor. | |
| void | Browse (TBrowser *b) |
| To browse the histograms. | |
| void | FillSWeightsHists (Int_t nbins=50) |
| The order of histograms in the array: | |
| void | FillXvarHists (Int_t nbins=100) |
| Fills the histograms of x variables (not weighted) with nbins. | |
| void | FillYpdfHists (Int_t nbins=100) |
| Fills the histograms of pdf-s of y variables with binning nbins. | |
| void | FillYvarHists (Int_t nbins=100) |
| Fill the histograms of y variables. | |
| Int_t | GetNevents () |
| Int_t | GetNspecies () |
| void | GetSWeights (Double_t *weights) |
| Returns the matrix of sweights. | |
| void | GetSWeights (TMatrixD &weights) |
| Returns the matrix of sweights. | |
| TH1D * | GetSWeightsHist (Int_t ixvar, Int_t ispecies, Int_t iyexcl=-1) |
| Returns the histogram of a variable, weighted with sWeights. | |
| TObjArray * | GetSWeightsHists () |
| Returns an array of all histograms of variables, weighted with sWeights. | |
| TString * | GetTreeExpression () |
| TString * | GetTreeName () |
| TString * | GetTreeSelection () |
| TH1D * | GetXvarHist (Int_t ixvar) |
| Returns the histogram of variable ixvar. | |
| TObjArray * | GetXvarHists () |
| Returns the array of histograms of x variables (not weighted). | |
| TH1D * | GetYpdfHist (Int_t iyvar, Int_t ispecies) |
| Returns the histogram of the pdf of variable iyvar for species #ispecies, binning nbins. | |
| TObjArray * | GetYpdfHists () |
| Returns the array of histograms of pdf's of y variables with binning nbins. | |
| TH1D * | GetYvarHist (Int_t iyvar) |
| Returns the histogram of variable iyvar.If histograms have not already been filled, they are filled with default binning 100. | |
| TObjArray * | GetYvarHists () |
| Returns the array of histograms of y variables. | |
| Bool_t | IsFolder () const |
| Returns kTRUE in case object contains browsable objects (like containers or lists of other objects). | |
| void | MakeSPlot (Option_t *option="v") |
| Calculates the sWeights. | |
| void | RefillHist (Int_t type, Int_t var, Int_t nbins, Double_t min, Double_t max, Int_t nspecies=-1) |
| The Fill...Hist() methods fill the histograms with the real limits on the variables This method allows to refill the specified histogram with user-set boundaries min and max. | |
| void | SetInitialNumbersOfSpecies (Int_t *numbers) |
| Set the initial number of events of each species - used as initial estimates in minuit. | |
| void | SetNEvents (Int_t ne) |
| void | SetNSpecies (Int_t ns) |
| void | SetNX (Int_t nx) |
| void | SetNY (Int_t ny) |
| void | SetTree (TTree *tree) |
| Set the input Tree. | |
| void | SetTreeSelection (const char *varexp="", const char *selection="", Long64_t firstentry=0) |
| Specifies the variables from the tree to be used for splot. | |
 Public Member Functions inherited from TObject Public Member Functions inherited from TObject | |
| TObject () | |
| TObject constructor. | |
| TObject (const TObject &object) | |
| TObject copy ctor. | |
| virtual | ~TObject () |
| TObject destructor. | |
| void | AbstractMethod (const char *method) const |
| Use this method to implement an "abstract" method that you don't want to leave purely abstract. | |
| virtual void | AppendPad (Option_t *option="") |
| Append graphics object to current pad. | |
| ULong_t | CheckedHash () |
| Check and record whether this class has a consistent Hash/RecursiveRemove setup (*) and then return the regular Hash value for this object. | |
| virtual const char * | ClassName () const |
| Returns name of class to which the object belongs. | |
| virtual void | Clear (Option_t *="") |
| virtual TObject * | Clone (const char *newname="") const |
| Make a clone of an object using the Streamer facility. | |
| virtual Int_t | Compare (const TObject *obj) const |
| Compare abstract method. | |
| virtual void | Copy (TObject &object) const |
| Copy this to obj. | |
| virtual void | Delete (Option_t *option="") |
| Delete this object. | |
| virtual Int_t | DistancetoPrimitive (Int_t px, Int_t py) |
| Computes distance from point (px,py) to the object. | |
| virtual void | Draw (Option_t *option="") |
| Default Draw method for all objects. | |
| virtual void | DrawClass () const |
| Draw class inheritance tree of the class to which this object belongs. | |
| virtual TObject * | DrawClone (Option_t *option="") const |
Draw a clone of this object in the current selected pad for instance with: gROOT->SetSelectedPad(gPad). | |
| virtual void | Dump () const |
| Dump contents of object on stdout. | |
| virtual void | Error (const char *method, const char *msgfmt,...) const |
| Issue error message. | |
| virtual void | Execute (const char *method, const char *params, Int_t *error=0) |
| Execute method on this object with the given parameter string, e.g. | |
| virtual void | Execute (TMethod *method, TObjArray *params, Int_t *error=0) |
| Execute method on this object with parameters stored in the TObjArray. | |
| virtual void | ExecuteEvent (Int_t event, Int_t px, Int_t py) |
| Execute action corresponding to an event at (px,py). | |
| virtual void | Fatal (const char *method, const char *msgfmt,...) const |
| Issue fatal error message. | |
| virtual TObject * | FindObject (const char *name) const |
| Must be redefined in derived classes. | |
| virtual TObject * | FindObject (const TObject *obj) const |
| Must be redefined in derived classes. | |
| virtual Option_t * | GetDrawOption () const |
| Get option used by the graphics system to draw this object. | |
| virtual const char * | GetIconName () const |
| Returns mime type name of object. | |
| virtual const char * | GetName () const |
| Returns name of object. | |
| virtual char * | GetObjectInfo (Int_t px, Int_t py) const |
| Returns string containing info about the object at position (px,py). | |
| virtual Option_t * | GetOption () const |
| virtual const char * | GetTitle () const |
| Returns title of object. | |
| virtual UInt_t | GetUniqueID () const |
| Return the unique object id. | |
| virtual Bool_t | HandleTimer (TTimer *timer) |
| Execute action in response of a timer timing out. | |
| virtual ULong_t | Hash () const |
| Return hash value for this object. | |
| Bool_t | HasInconsistentHash () const |
| Return true is the type of this object is known to have an inconsistent setup for Hash and RecursiveRemove (i.e. | |
| virtual void | Info (const char *method, const char *msgfmt,...) const |
| Issue info message. | |
| virtual Bool_t | InheritsFrom (const char *classname) const |
| Returns kTRUE if object inherits from class "classname". | |
| virtual Bool_t | InheritsFrom (const TClass *cl) const |
| Returns kTRUE if object inherits from TClass cl. | |
| virtual void | Inspect () const |
| Dump contents of this object in a graphics canvas. | |
| void | InvertBit (UInt_t f) |
| virtual Bool_t | IsEqual (const TObject *obj) const |
| Default equal comparison (objects are equal if they have the same address in memory). | |
| R__ALWAYS_INLINE Bool_t | IsOnHeap () const |
| virtual Bool_t | IsSortable () const |
| R__ALWAYS_INLINE Bool_t | IsZombie () const |
| virtual void | ls (Option_t *option="") const |
| The ls function lists the contents of a class on stdout. | |
| void | MayNotUse (const char *method) const |
| Use this method to signal that a method (defined in a base class) may not be called in a derived class (in principle against good design since a child class should not provide less functionality than its parent, however, sometimes it is necessary). | |
| virtual Bool_t | Notify () |
| This method must be overridden to handle object notification. | |
| void | Obsolete (const char *method, const char *asOfVers, const char *removedFromVers) const |
| Use this method to declare a method obsolete. | |
| void | operator delete (void *ptr) |
| Operator delete. | |
| void | operator delete[] (void *ptr) |
| Operator delete []. | |
| void * | operator new (size_t sz) |
| void * | operator new (size_t sz, void *vp) |
| void * | operator new[] (size_t sz) |
| void * | operator new[] (size_t sz, void *vp) |
| TObject & | operator= (const TObject &rhs) |
| TObject assignment operator. | |
| virtual void | Paint (Option_t *option="") |
| This method must be overridden if a class wants to paint itself. | |
| virtual void | Pop () |
| Pop on object drawn in a pad to the top of the display list. | |
| virtual void | Print (Option_t *option="") const |
| This method must be overridden when a class wants to print itself. | |
| virtual Int_t | Read (const char *name) |
| Read contents of object with specified name from the current directory. | |
| virtual void | RecursiveRemove (TObject *obj) |
| Recursively remove this object from a list. | |
| void | ResetBit (UInt_t f) |
| virtual void | SaveAs (const char *filename="", Option_t *option="") const |
| Save this object in the file specified by filename. | |
| virtual void | SavePrimitive (std::ostream &out, Option_t *option="") |
| Save a primitive as a C++ statement(s) on output stream "out". | |
| void | SetBit (UInt_t f) |
| void | SetBit (UInt_t f, Bool_t set) |
| Set or unset the user status bits as specified in f. | |
| virtual void | SetDrawOption (Option_t *option="") |
| Set drawing option for object. | |
| virtual void | SetUniqueID (UInt_t uid) |
| Set the unique object id. | |
| virtual void | SysError (const char *method, const char *msgfmt,...) const |
| Issue system error message. | |
| R__ALWAYS_INLINE Bool_t | TestBit (UInt_t f) const |
| Int_t | TestBits (UInt_t f) const |
| virtual void | UseCurrentStyle () |
| Set current style settings in this object This function is called when either TCanvas::UseCurrentStyle or TROOT::ForceStyle have been invoked. | |
| virtual void | Warning (const char *method, const char *msgfmt,...) const |
| Issue warning message. | |
| virtual Int_t | Write (const char *name=0, Int_t option=0, Int_t bufsize=0) |
| Write this object to the current directory. | |
| virtual Int_t | Write (const char *name=0, Int_t option=0, Int_t bufsize=0) const |
| Write this object to the current directory. | |
Protected Member Functions | |
| void | SPlots (Double_t *covmat, Int_t i_excl) |
| Computes the sWeights from the covariance matrix. | |
 Protected Member Functions inherited from TObject Protected Member Functions inherited from TObject | |
| virtual void | DoError (int level, const char *location, const char *fmt, va_list va) const |
| Interface to ErrorHandler (protected). | |
| void | MakeZombie () |
Additional Inherited Members | |
 Public Types inherited from TObject Public Types inherited from TObject | |
| enum | { kIsOnHeap = 0x01000000 , kNotDeleted = 0x02000000 , kZombie = 0x04000000 , kInconsistent = 0x08000000 , kBitMask = 0x00ffffff } |
| enum | { kSingleKey = BIT(0) , kOverwrite = BIT(1) , kWriteDelete = BIT(2) } |
| enum | EDeprecatedStatusBits { kObjInCanvas = BIT(3) } |
| enum | EStatusBits { kCanDelete = BIT(0) , kMustCleanup = BIT(3) , kIsReferenced = BIT(4) , kHasUUID = BIT(5) , kCannotPick = BIT(6) , kNoContextMenu = BIT(8) , kInvalidObject = BIT(13) } |
 Static Public Member Functions inherited from TObject Static Public Member Functions inherited from TObject | |
| static Long_t | GetDtorOnly () |
| Return destructor only flag. | |
| static Bool_t | GetObjectStat () |
| Get status of object stat flag. | |
| static void | SetDtorOnly (void *obj) |
| Set destructor only flag. | |
| static void | SetObjectStat (Bool_t stat) |
| Turn on/off tracking of objects in the TObjectTable. | |
 Protected Types inherited from TObject Protected Types inherited from TObject | |
| enum | { kOnlyPrepStep = BIT(3) } |
#include <TSPlot.h>
| TSPlot::TSPlot | ( | ) |
default constructor (used by I/O only)
Definition at line 296 of file TSPlot.cxx.
Normal TSPlot constructor.
Definition at line 317 of file TSPlot.cxx.
|
virtual |
Destructor.
Definition at line 339 of file TSPlot.cxx.
The order of histograms in the array:
x0_species0, x0_species1,..., x1_species0, x1_species1,..., y0_species0, y0_species1,...
If the histograms have already been filled with a different binning, they are refilled and all histograms are deleted
Definition at line 698 of file TSPlot.cxx.
Fills the histograms of x variables (not weighted) with nbins.
Definition at line 528 of file TSPlot.cxx.
Fills the histograms of pdf-s of y variables with binning nbins.
Definition at line 638 of file TSPlot.cxx.
Fill the histograms of y variables.
Definition at line 585 of file TSPlot.cxx.
Returns the matrix of sweights.
It is assumed that the array passed in the argurment is big enough
Definition at line 516 of file TSPlot.cxx.
Returns the matrix of sweights.
Definition at line 505 of file TSPlot.cxx.
Returns the histogram of a variable, weighted with sWeights.
Definition at line 832 of file TSPlot.cxx.
| TObjArray * TSPlot::GetSWeightsHists | ( | ) |
Returns an array of all histograms of variables, weighted with sWeights.
If histograms have not been already filled, they are filled with default binning 50 The order of histograms in the array:
x0_species0, x0_species1,..., x1_species0, x1_species1,..., y0_species0, y0_species1,...
Definition at line 746 of file TSPlot.cxx.
Returns the histogram of variable ixvar.
If histograms have not already been filled, they are filled with default binning 100.
Definition at line 571 of file TSPlot.cxx.
| TObjArray * TSPlot::GetXvarHists | ( | ) |
Returns the array of histograms of x variables (not weighted).
If histograms have not already been filled, they are filled with default binning 100.
Definition at line 556 of file TSPlot.cxx.
Returns the histogram of the pdf of variable iyvar for species #ispecies, binning nbins.
If histograms have not already been filled, they are filled with default binning 100.
Definition at line 681 of file TSPlot.cxx.
| TObjArray * TSPlot::GetYpdfHists | ( | ) |
Returns the array of histograms of pdf's of y variables with binning nbins.
If histograms have not already been filled, they are filled with default binning 100.
Definition at line 667 of file TSPlot.cxx.
Returns the histogram of variable iyvar.If histograms have not already been filled, they are filled with default binning 100.
Definition at line 625 of file TSPlot.cxx.
| TObjArray * TSPlot::GetYvarHists | ( | ) |
Returns the array of histograms of y variables.
If histograms have not already been filled, they are filled with default binning 100.
Definition at line 611 of file TSPlot.cxx.
|
inlinevirtual |
Calculates the sWeights.
The option controls the print level
Definition at line 404 of file TSPlot.cxx.
| void TSPlot::RefillHist | ( | Int_t | type, |
| Int_t | nvar, | ||
| Int_t | nbins, | ||
| Double_t | min, | ||
| Double_t | max, | ||
| Int_t | nspecies = -1 |
||
| ) |
The Fill...Hist() methods fill the histograms with the real limits on the variables This method allows to refill the specified histogram with user-set boundaries min and max.
Parameters:
Definition at line 767 of file TSPlot.cxx.
Set the initial number of events of each species - used as initial estimates in minuit.
Definition at line 388 of file TSPlot.cxx.
Set the input Tree.
Definition at line 850 of file TSPlot.cxx.
| void TSPlot::SetTreeSelection | ( | const char * | varexp = "", |
| const char * | selection = "", |
||
| Long64_t | firstentry = 0 |
||
| ) |
Specifies the variables from the tree to be used for splot.
Variables fNx, fNy, fNSpecies and fNEvents should already be set!
In the 1st parameter it is assumed that first fNx variables are x(control variables), then fNy y variables (discriminating variables), then fNy*fNSpecies ypdf variables (probability distribution functions of discriminating variables for different species). The order of pdfs should be: species0_y0, species0_y1,... species1_y0, species1_y1,...species[fNSpecies-1]_y0... The 2nd parameter allows to make a cut TTree::Draw method description contains more details on specifying expression and selection
Definition at line 868 of file TSPlot.cxx.
Computes the sWeights from the covariance matrix.
Definition at line 484 of file TSPlot.cxx.