
class TMVA::MethodLikelihood: public TMVA::MethodBase
Likelihood analysis ("non-parametric approach")
Also implemented is a "diagonalized likelihood approach", which improves over the uncorrelated likelihood ansatz by transforming linearly the input variables into a diagonal space, using the square-root of the covariance matrix
The method of maximum likelihood is the most straightforward, and
certainly among the most elegant multivariate analyser approaches.
We define the likelihood ratio, RL, for event
i, by:

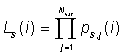
Note that in TMVA the output of the likelihood ratio is transformed
by

The biggest drawback of the Likelihood approach is that it assumes
that the discriminant variables are uncorrelated. If it were the case,
it can be proven that the discrimination obtained by the above likelihood
ratio is optimal, ie, no other method can beat it. However, in most
practical applications of MVAs correlations are present.
Linear correlations, measured from the training sample, can be taken
into account in a straightforward manner through the square-root
of the covariance matrix. The square-root of a matrix
C is the matrix C′ that multiplied with itself
yields C: C=C′C′. We compute the
square-root matrix (SQM) by means of diagonalising (D) the
covariance matrix:

The above diagonalisation is complete for linearly correlated, Gaussian distributed variables only. In real-world examples this is not often the case, so that only little additional information may be recovered by the diagonalisation procedure. In these cases, non-linear methods must be applied.
Function Members (Methods)
| virtual void | DeclareOptions() |
| virtual void | Init() |
| virtual void | ProcessOptions() |
| Double_t | TransformLikelihoodOutput(Double_t ps, Double_t pb) const |
Data Members
| enum TMVA::MethodBase::EWeightFileType { | kROOT | |
| kTEXT | ||
| }; | ||
| enum TMVA::MethodBase::ECutOrientation { | kNegative | |
| kPositive | ||
| }; | ||
| enum TObject::EStatusBits { | kCanDelete | |
| kMustCleanup | ||
| kObjInCanvas | ||
| kIsReferenced | ||
| kHasUUID | ||
| kCannotPick | ||
| kNoContextMenu | ||
| kInvalidObject | ||
| }; | ||
| enum TObject::[unnamed] { | kIsOnHeap | |
| kNotDeleted | ||
| kZombie | ||
| kBitMask | ||
| kSingleKey | ||
| kOverwrite | ||
| kWriteDelete | ||
| }; |
| Bool_t | TMVA::MethodBase::fSetupCompleted | is method setup |
| const TMVA::Event* | TMVA::MethodBase::fTmpEvent | ! temporary event when testing on a different DataSet than the own one |
| TMVA::Types::EAnalysisType | TMVA::MethodBase::fAnalysisType | method-mode : true --> regression, false --> classification |
| UInt_t | TMVA::MethodBase::fBackgroundClass | index of the Background-class |
| vector<TString>* | TMVA::MethodBase::fInputVars | vector of input variables used in MVA |
| Int_t | TMVA::MethodBase::fNbins | number of bins in representative histograms |
| Int_t | TMVA::MethodBase::fNbinsH | number of bins in evaluation histograms |
| TMVA::Ranking* | TMVA::MethodBase::fRanking | pointer to ranking object (created by derived classifiers) |
| vector<Float_t>* | TMVA::MethodBase::fRegressionReturnVal | holds the return-value for the regression |
| UInt_t | TMVA::MethodBase::fSignalClass | index of the Signal-class |
| Int_t | fAverageEvtPerBin | average events per bin; used to calculate fNbins |
| Int_t* | fAverageEvtPerBinVarB | average events per bin; used to calculate fNbins |
| Int_t* | fAverageEvtPerBinVarS | average events per bin; used to calculate fNbins |
| TString | fBorderMethodString | the method to take care about "border" effects (string) |
| TMVA::PDF* | fDefaultPDFLik | pdf that contains default definitions |
| Int_t | fDropVariable | for ranking test |
| Double_t | fEpsilon | minimum number of likelihood (to avoid zero) |
| vector<TH1*>* | fHistBgd | background PDFs (histograms) |
| vector<TH1*>* | fHistBgd_smooth | background PDFs (smoothed histograms) |
| vector<TH1*>* | fHistSig | signal PDFs (histograms) |
| vector<TH1*>* | fHistSig_smooth | signal PDFs (smoothed histograms) |
| TString* | fInterpolateString | which interpolation method used for reference histograms (individual for each variable) |
| Float_t | fKDEfineFactor | fine tuning factor for Adaptive KDE |
| TString | fKDEiterString | Number of iterations (string) |
| TString | fKDEtypeString | Kernel type to use for KDE (string) |
| Int_t | fNsmooth | number of smooth passes |
| Int_t* | fNsmoothVarB | number of smooth passes |
| Int_t* | fNsmoothVarS | number of smooth passes |
| vector<PDF*>* | fPDFBgd | list of PDFs (background) |
| vector<PDF*>* | fPDFSig | list of PDFs (signal) |
| Bool_t | fTransformLikelihoodOutput | likelihood output is sigmoid-transformed |
Class Charts
Function documentation
standard constructor
construct likelihood references from file
FDA can handle classification with 2 classes
define the options (their key words) that can be set in the option string TransformOutput <bool> transform (often strongly peaked) likelihood output through sigmoid inversion
create reference distributions (PDFs) from signal and background events: fill histograms and smooth them; if decorrelation is required, compute corresponding square-root matrices the reference histograms require the correct boundaries. Since in Likelihood classification the transformations are applied using both classes, also the corresponding boundaries need to take this into account
returns the likelihood estimator for signal fill a new Likelihood branch into the testTree
returns transformed or non-transformed output
read weight info from file nothing to do for this method
write specific header of the classifier (mostly include files)
get help message text
typical length of text line:
"|--------------------------------------------------------------|"