The macro creates a simple signal model and two background models, which are added to a RooWorkspace. The macro creates a toy dataset, and then uses a RooStats ProfileLikleihoodCalculator to do a hypothesis test of the background-only and signal+background hypotheses. In this example, shape uncertainties are not taken into account, but normalization uncertainties are.
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooAddPdf::model
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooGaussian::sigModel
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooRealVar::invMass
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooRealVar::mH
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooRealVar::sigma1
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooProduct::fsig
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooRealVar::mu
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooRealVar::ratioSigEff
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooRealVar::fsigExpected
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooGaussian::zjjModel
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooRealVar::mZ
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooRealVar::sigma1_z
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooRealVar::fzjj
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooChebychev::qcdModel
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooRealVar::a0
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooRealVar::a1
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooRealVar::a2
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing dataset modelData
[#1] INFO:ObjectHandling -- RooWorkSpace::import(myWS) changing name of dataset from modelData to data
[#1] INFO:InputArguments -- The deprecated RooFit::CloneData(1) option passed to createNLL() is ignored.
[#1] INFO:Fitting -- RooAbsPdf::fitTo(model) fixing normalization set for coefficient determination to observables in data
[#1] INFO:Fitting -- using generic CPU library compiled with no vectorizations
[#1] INFO:Fitting -- Creation of NLL object took 531.052 μs
[#0] PROGRESS:Minimization -- ProfileLikelihoodCalcultor::DoGLobalFit - find MLE
[#1] INFO:Fitting -- RooAddition::defaultErrorLevel(nll_model_data) Summation contains a RooNLLVar, using its error level
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: activating const optimization
[#0] PROGRESS:Minimization -- ProfileLikelihoodCalcultor::DoMinimizeNLL - using Minuit2 / with strategy 1
[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
[#1] INFO:Minimization --
RooFitResult: minimized FCN value: 717.039, estimated distance to minimum: 4.45135e-10
covariance matrix quality: Full, accurate covariance matrix
Status : MINIMIZE=0
Floating Parameter FinalValue +/- Error
-------------------- --------------------------
fzjj 3.1152e-01 +/- 5.03e-02
mu 1.0968e+00 +/- 3.03e-01
[#0] PROGRESS:Minimization -- ProfileLikelihoodCalcultor::GetHypoTest - do conditional fit
[#1] INFO:Fitting -- RooAddition::defaultErrorLevel(nll_model_data) Summation contains a RooNLLVar, using its error level
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: activating const optimization
[#0] PROGRESS:Minimization -- ProfileLikelihoodCalcultor::DoMinimizeNLL - using Minuit2 / with strategy 1
[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
[#1] INFO:Minimization --
RooFitResult: minimized FCN value: 723.97, estimated distance to minimum: 1.04934e-09
covariance matrix quality: Full, accurate covariance matrix
Status : MINIMIZE=0
Floating Parameter FinalValue +/- Error
-------------------- --------------------------
fzjj 2.6213e-01 +/- 5.18e-02
-------------------------------------------------
The p-value for the null is 9.83108e-05
Corresponding to a significance of 3.72332
-------------------------------------------------
[#1] INFO:Fitting -- RooAbsPdf::fitTo(model) fixing normalization set for coefficient determination to observables in data
[#1] INFO:Fitting -- Creation of NLL object took 120.699 μs
[#1] INFO:Fitting -- RooAddition::defaultErrorLevel(nll_model_data) Summation contains a RooNLLVar, using its error level
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: activating const optimization
[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: deactivating const optimization
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model) directly selected PDF components: (sigModel)
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model) indirectly selected PDF components: ()
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model) directly selected PDF components: (zjjModel)
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model) indirectly selected PDF components: ()
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model) directly selected PDF components: (qcdModel)
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model) indirectly selected PDF components: ()
[#1] INFO:Fitting -- RooAbsPdf::fitTo(model) fixing normalization set for coefficient determination to observables in data
[#1] INFO:Fitting -- Creation of NLL object took 97.791 μs
[#1] INFO:Fitting -- RooAddition::defaultErrorLevel(nll_model_data) Summation contains a RooNLLVar, using its error level
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: activating const optimization
[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: deactivating const optimization
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model) directly selected PDF components: (zjjModel)
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model) indirectly selected PDF components: ()
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model) directly selected PDF components: (qcdModel)
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model) indirectly selected PDF components: ()
#include <string>
void rs102_hypotestwithshapes()
{
AddModel(wspace);
AddData(wspace);
delete wspace;
}
{
RooRealVar fzjj(
"fzjj",
"fraction of zjj background events", .4, 0., 1);
RooRealVar mu(
"mu",
"signal strength in units of SM expectation", 1, 0., 2);
RooRealVar ratioSigEff(
"ratioSigEff",
"ratio of signal efficiency to nominal signal efficiency", 1., 0., 2);
}
{
}
{
plc.SetData(*(
wks->data(
"data")));
cout << "-------------------------------------------------" << endl;
cout <<
"The p-value for the null is " <<
htr->NullPValue() << endl;
cout <<
"Corresponding to a significance of " <<
htr->Significance() << endl;
cout << "-------------------------------------------------\n\n" << endl;
}
{
frame->
SetTitle(
"An example fit to the signal + background model");
xframe2->SetTitle(
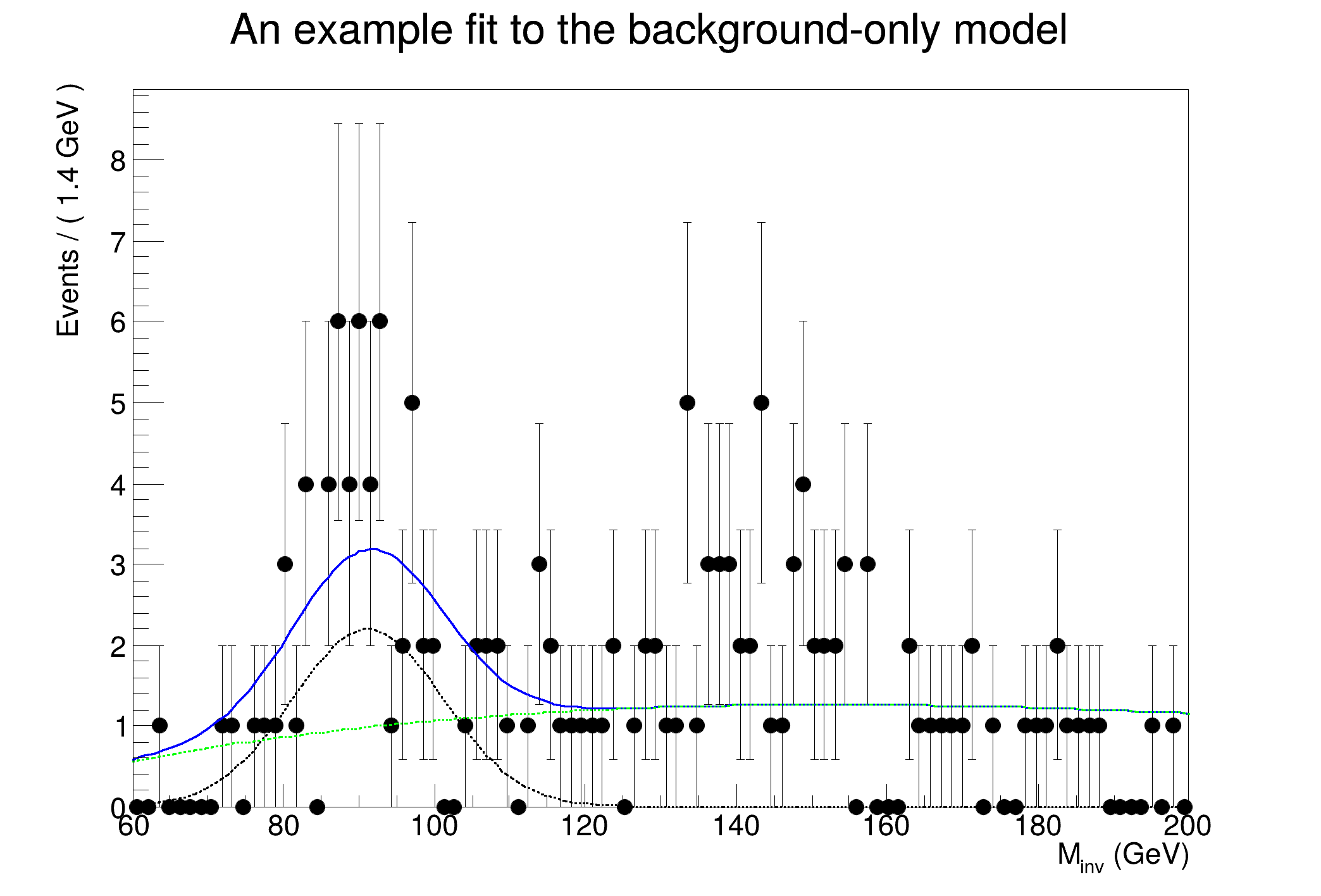
"An example fit to the background-only model");
}
int Int_t
Signed integer 4 bytes (int)
double Double_t
Double 8 bytes.
ROOT::Detail::TRangeCast< T, true > TRangeDynCast
TRangeDynCast is an adapter class that allows the typed iteration through a TCollection.
Option_t Option_t TPoint TPoint const char GetTextMagnitude GetFillStyle GetLineColor GetLineWidth GetMarkerStyle GetTextAlign GetTextColor GetTextSize void data
Abstract base class for binned and unbinned datasets.
Abstract interface for all probability density functions.
RooPlot * plotOn(RooPlot *frame, const RooCmdArg &arg1={}, const RooCmdArg &arg2={}, const RooCmdArg &arg3={}, const RooCmdArg &arg4={}, const RooCmdArg &arg5={}, const RooCmdArg &arg6={}, const RooCmdArg &arg7={}, const RooCmdArg &arg8={}, const RooCmdArg &arg9={}, const RooCmdArg &arg10={}) const override
Helper calling plotOn(RooPlot*, RooLinkedList&) const.
RooFit::OwningPtr< RooFitResult > fitTo(RooAbsData &data, CmdArgs_t const &... cmdArgs)
Fit PDF to given dataset.
RooFit::OwningPtr< RooDataSet > generate(const RooArgSet &whatVars, Int_t nEvents, const RooCmdArg &arg1, const RooCmdArg &arg2={}, const RooCmdArg &arg3={}, const RooCmdArg &arg4={}, const RooCmdArg &arg5={})
See RooAbsPdf::generate(const RooArgSet&,const RooCmdArg&,const RooCmdArg&,const RooCmdArg&,...
void setConstant(bool value=true)
Efficient implementation of a sum of PDFs of the form.
RooArgList is a container object that can hold multiple RooAbsArg objects.
RooArgSet is a container object that can hold multiple RooAbsArg objects.
Chebychev polynomial p.d.f.
Plot frame and a container for graphics objects within that frame.
void SetTitle(const char *name) override
Set the title of the RooPlot to 'title'.
void Draw(Option_t *options=nullptr) override
Draw this plot and all of the elements it contains.
Represents the product of a given set of RooAbsReal objects.
Variable that can be changed from the outside.
void setVal(double value) override
Set value of variable to 'value'.
HypoTestResult is a base class for results from hypothesis tests.
< A class that holds configuration information for a model using a workspace as a store
void SetWorkspace(RooWorkspace &ws)
void SetPdf(const RooAbsPdf &pdf)
Set the Pdf, add to the workspace if not already there.
The ProfileLikelihoodCalculator is a concrete implementation of CombinedCalculator (the interface cla...
Persistable container for RooFit projects.
RooCmdArg Rename(const char *suffix)
RooCmdArg Hesse(bool flag=true)
RooCmdArg Save(bool flag=true)
RooCmdArg Minos(bool flag=true)
RooCmdArg PrintLevel(Int_t code)
RooCmdArg DataError(Int_t)
RooCmdArg Components(Args_t &&... argsOrArgSet)
RooCmdArg LineColor(TColorNumber color)
RooCmdArg LineStyle(Style_t style)
The namespace RooFit contains mostly switches that change the behaviour of functions of PDFs (or othe...
Namespace for the RooStats classes.