ROOT's RDataFrame offers a modern, high-level interface for analysis of data stored in TTree , CSV and other data formats, in C++ or Python.
In addition, multi-threading and other low-level optimisations allow users to exploit all the resources available on their machines completely transparently.
Skip to the class reference or keep reading for the user guide.
In a nutshell:
Calculations are expressed in terms of a type-safe functional chain of actions and transformations, RDataFrame takes care of their execution. The implementation automatically puts in place several low level optimisations such as multi-thread parallelization and caching.
You can directly see RDataFrame in action in our tutorials, in C++ or Python.
These are the operations which can be performed with RDataFrame.
Transformations are a way to manipulate the data.
| Transformation | Description |
|---|---|
| Alias() | Introduce an alias for a particular column name. |
| Define() | Create a new column in the dataset. Example usages include adding a column that contains the invariant mass of a particle, or a selection of elements of an array (e.g. only the pts of "good" muons). |
| DefinePerSample() | Define a new column that is updated when the input sample changes, e.g. when switching tree being processed in a chain. |
| DefineSlot() | Same as Define(), but the user-defined function must take an extra unsigned int slot as its first parameter. slot will take a different value, 0 to nThreads - 1, for each thread of execution. This is meant as a helper in writing thread-safe Define() transformation when using RDataFrame after ROOT::EnableImplicitMT(). DefineSlot() works just as well with single-thread execution: in that case slot will always be 0. |
| DefineSlotEntry() | Same as DefineSlot(), but the entry number is passed in addition to the slot number. This is meant as a helper in case the expression depends on the entry number. For details about entry numbers in multi-threaded runs, see here. |
| Filter() | Filter rows based on user-defined conditions. |
| Range() | Filter rows based on entry number (single-thread only). |
| Redefine() | Overwrite the value and/or type of an existing column. See Define() for more information. |
| RedefineSlot() | Overwrite the value and/or type of an existing column. See DefineSlot() for more information. |
| RedefineSlotEntry() | Overwrite the value and/or type of an existing column. See DefineSlotEntry() for more information. |
| Vary() | Register systematic variations for an existing column. Varied results are then extracted via VariationsFor(). |
Actions aggregate data into a result. Each one is described in more detail in the reference guide.
In the following, whenever we say an action "returns" something, we always mean it returns a smart pointer to it. Actions only act on events that pass all preceding filters.
Lazy actions only trigger the event loop when one of the results is accessed for the first time, making it easy to produce many different results in one event loop. Instant actions trigger the event loop instantly.
| Lazy action | Description |
|---|---|
| Aggregate() | Execute a user-defined accumulation operation on the processed column values. |
| Book() | Book execution of a custom action using a user-defined helper object. |
| Cache() | Cache column values in memory. Custom columns can be cached as well, filtered entries are not cached. Users can specify which columns to save (default is all). |
| Count() | Return the number of events processed. Useful e.g. to get a quick count of the number of events passing a Filter. |
| Display() | Provides a printable representation of the dataset contents. The method returns a ROOT::RDF::RDisplay() instance which can print a tabular representation of the data or return it as a string. |
| Fill() | Fill a user-defined object with the values of the specified columns, as if by calling Obj.Fill(col1, col2, ...). |
| Graph() | Fills a TGraph with the two columns provided. If multi-threading is enabled, the order of the points may not be the one expected, it is therefore suggested to sort if before drawing. |
| GraphAsymmErrors() | Fills a TGraphAsymmErrors. If multi-threading is enabled, the order of the points may not be the one expected, it is therefore suggested to sort if before drawing. |
| Histo1D(), Histo2D(), Histo3D() | Fill a one-, two-, three-dimensional histogram with the processed column values. |
| HistoND() | Fill an N-dimensional histogram with the processed column values. |
| Max() | Return the maximum of processed column values. If the type of the column is inferred, the return type is double, the type of the column otherwise. |
| Mean() | Return the mean of processed column values. |
| Min() | Return the minimum of processed column values. If the type of the column is inferred, the return type is double, the type of the column otherwise. |
| Profile1D(), Profile2D() | Fill a one- or two-dimensional profile with the column values that passed all filters. |
| Reduce() | Reduce (e.g. sum, merge) entries using the function (lambda, functor...) passed as argument. The function must have signature T(T,T) where T is the type of the column. Return the final result of the reduction operation. An optional parameter allows initialization of the result object to non-default values. |
| Report() | Obtain statistics on how many entries have been accepted and rejected by the filters. See the section on named filters for a more detailed explanation. The method returns a ROOT::RDF::RCutFlowReport instance which can be queried programmatically to get information about the effects of the individual cuts. |
| Stats() | Return a TStatistic object filled with the input columns. |
| StdDev() | Return the unbiased standard deviation of the processed column values. |
| Sum() | Return the sum of the values in the column. If the type of the column is inferred, the return type is double, the type of the column otherwise. |
| Take() | Extract a column from the dataset as a collection of values, e.g. a std::vector<float> for a column of type float. |
| Instant action | Description |
|---|---|
| Foreach() | Execute a user-defined function on each entry. Users are responsible for the thread-safety of this callable when executing with implicit multi-threading enabled. |
| ForeachSlot() | Same as Foreach(), but the user-defined function must take an extra unsigned int slot as its first parameter. slot will take a different value, 0 to nThreads - 1, for each thread of execution. This is meant as a helper in writing thread-safe Foreach() actions when using RDataFrame after ROOT::EnableImplicitMT(). ForeachSlot() works just as well with single-thread execution: in that case slot will always be 0. |
| Snapshot() | Write the processed dataset to disk, in a new TTree and TFile. Custom columns can be saved as well, filtered entries are not saved. Users can specify which columns to save (default is all). Snapshot, by default, overwrites the output file if it already exists. Snapshot() can be made lazy setting the appropriate flag in the snapshot options. |
These operations do not modify the dataframe or book computations but simply return information on the RDataFrame object.
| Operation | Description |
|---|---|
| Describe() | Get useful information describing the dataframe, e.g. columns and their types. |
| GetColumnNames() | Get the names of all the available columns of the dataset. |
| GetColumnType() | Return the type of a given column as a string. |
| GetColumnTypeNamesList() | Return the list of type names of columns in the dataset. |
| GetDefinedColumnNames() | Get the names of all the defined columns. |
| GetFilterNames() | Return the names of all filters in the computation graph. |
| GetNRuns() | Return the number of event loops run by this RDataFrame instance so far. |
| GetNSlots() | Return the number of processing slots that RDataFrame will use during the event loop (i.e. the concurrency level). |
| SaveGraph() | Store the computation graph of an RDataFrame in DOT format (graphviz) for easy inspection. See the relevant section for details. |
Users define their analysis as a sequence of operations to be performed on the dataframe object; the framework takes care of the management of the loop over entries as well as low-level details such as I/O and parallelization. RDataFrame provides methods to perform most common operations required by ROOT analyses; at the same time, users can just as easily specify custom code that will be executed in the event loop.
RDataFrame is built with a modular and flexible workflow in mind, summarised as follows:
Make sure to book all transformations and actions before you access the contents of any of the results. This lets RDataFrame accumulate work and then produce all results at the same time, upon first access to any of them.
The following table shows how analyses based on TTreeReader and TTree::Draw() translate to RDataFrame. Follow the crash course to discover more idiomatic and flexible ways to express analyses with RDataFrame.
| TTreeReader | ROOT::RDataFrame |
while(reader.Next()) {
}
An interface for reading values stored in ROOT columnar datasets. Definition TTreeReaderValue.h:146 A simple, robust and fast interface to read values from ROOT columnar datasets such as TTree,... Definition TTreeReader.h:44 | RInterface< RDFDetail::RFilter< F, Proxied >, DS_t > Filter(F f, const ColumnNames_t &columns={}, std::string_view name="") Append a filter to the call graph. Definition RInterface.hxx:216 |
| TTree::Draw | ROOT::RDataFrame |
h->Draw()
| |
df.Filter("event == 1").Histo1D("jet_eta", "weight");
// or the fully compiled version:
df.Filter([] (ULong64_t e) { return e == 1; }, {"event"}).Histo1D<RVec<float>>("jet_eta", "weight");
RResultPtr<::TH1D > Histo1D(const TH1DModel &model={"", "", 128u, 0., 0.}, std::string_view vName="") Fill and return a one-dimensional histogram with the values of a column (lazy action). Definition RInterface.hxx:1479 | |
// object selection: for each event, fill histogram with array of selected pts
| // with RDF, arrays are read as ROOT::VecOps::RVec objects
df.Define("good_pt", "Muon_pt[Muon_pt > 100]").Histo1D("good_pt")
|
All snippets of code presented in the crash course can be executed in the ROOT interpreter. Simply precede them with
which is omitted for brevity. The terms "column" and "branch" are used interchangeably.
RDataFrame's constructor is where the user specifies the dataset and, optionally, a default set of columns that operations should work with. Here are the most common methods to construct an RDataFrame object:
Additionally, users can construct an RDataFrame with no data source by passing an integer number. This is the number of rows that will be generated by this RDataFrame.
This is useful to generate simple datasets on the fly: the contents of each event can be specified with Define() (explained below). For example, we have used this method to generate Pythia events and write them to disk in parallel (with the Snapshot action).
For data sources other than TTrees and TChains, RDataFrame objects are constructed using ad-hoc factory functions (see e.g. FromCSV(), FromSqlite(), FromArrow()):
Let's now tackle a very common task, filling a histogram:
The first line creates an RDataFrame associated to the TTree "myTree". This tree has a branch named "MET".
Histo1D() is an action; it returns a smart pointer (a ROOT::RDF::RResultPtr, to be precise) to a TH1D histogram filled with the MET of all events. If the quantity stored in the column is a collection (e.g. a vector or an array), the histogram is filled with all vector elements for each event.
You can use the objects returned by actions as if they were pointers to the desired results. There are many other possible actions, and all their results are wrapped in smart pointers; we'll see why in a minute.
Let's say we want to cut over the value of branch "MET" and count how many events pass this cut. This is one way to do it:
The filter string (which must contain a valid C++ expression) is applied to the specified columns for each event; the name and types of the columns are inferred automatically. The string expression is required to return a bool which signals whether the event passes the filter (true) or not (false).
You can think of your data as "flowing" through the chain of calls, being transformed, filtered and finally used to perform actions. Multiple Filter() calls can be chained one after another.
Using string filters is nice for simple things, but they are limited to specifying the equivalent of a single return statement or the body of a lambda, so it's cumbersome to use strings with more complex filters. They also add a small runtime overhead, as ROOT needs to just-in-time compile the string into C++ code. When more freedom is required or runtime performance is very important, a C++ callable can be specified instead (a lambda in the following snippet, but it can be any kind of function or even a functor class), together with a list of column names. This snippet is analogous to the one above:
An example of a more complex filter expressed as a string containing C++ code is shown below
The code snippet above defines a column p that is a fixed-size array using the component column names and then filters on its magnitude by looping over its elements. It must be noted that the usage of strings to define columns like the one above is currently the only possibility when using PyROOT. When writing expressions as such, only constants and data coming from other columns in the dataset can be involved in the code passed as a string. Local variables and functions cannot be used, since the interpreter will not know how to find them. When capturing local state is necessary, it must first be declared to the ROOT C++ interpreter.
More information on filters and how to use them to automatically generate cutflow reports can be found below.
Let's now consider the case in which "myTree" contains two quantities "x" and "y", but our analysis relies on a derived quantity z = sqrt(x*x + y*y). Using the Define() transformation, we can create a new column in the dataset containing the variable "z":
Define() creates the variable "z" by applying sqrtSum to "x" and "y". Later in the chain of calls we refer to variables created with Define() as if they were actual tree branches/columns, but they are evaluated on demand, at most once per event. As with filters, Define() calls can be chained with other transformations to create multiple custom columns. Define() and Filter() transformations can be concatenated and intermixed at will.
As with filters, it is possible to specify new columns as string expressions. This snippet is analogous to the one above:
Again the names of the columns used in the expression and their types are inferred automatically. The string must be valid C++ and it is just-in-time compiled. The process has a small runtime overhead and like with filters it is currently the only possible approach when using PyROOT.
Previously, when showing the different ways an RDataFrame can be created, we showed a constructor that takes a number of entries as a parameter. In the following example we show how to combine such an "empty" RDataFrame with Define() transformations to create a dataset on the fly. We then save the generated data on disk using the Snapshot() action.
This example is slightly more advanced than what we have seen so far. First, it makes use of lambda captures (a simple way to make external variables available inside the body of C++ lambdas) to act on the same variable x from both Define() transformations. Second, we have stored the transformed dataframe in a variable. This is always possible, since at each point of the transformation chain users can store the status of the dataframe for further use (more on this below).
You can read more about defining new columns here.
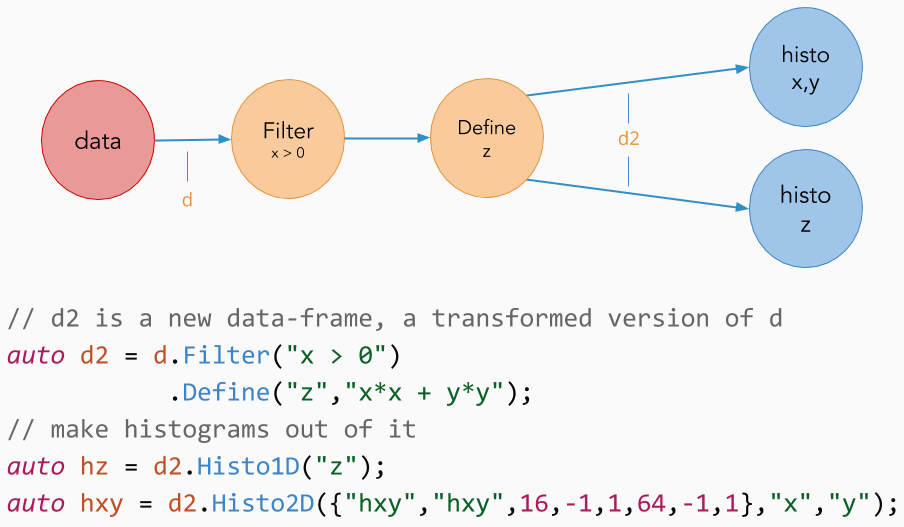
It is sometimes necessary to limit the processing of the dataset to a range of entries. For this reason, the RDataFrame offers the concept of ranges as a node of the RDataFrame chain of transformations; this means that filters, columns and actions can be concatenated to and intermixed with Range()s. If a range is specified after a filter, the range will act exclusively on the entries passing the filter – it will not even count the other entries! The same goes for a Range() hanging from another Range(). Here are some commented examples:
Note that ranges are not available when multi-threading is enabled. More information on ranges is available here.
As a final example let us apply two different cuts on branch "MET" and fill two different histograms with the "pt_v" of the filtered events. By now, you should be able to easily understand what is happening:
RDataFrame executes all above actions by running the event-loop only once. The trick is that actions are not executed at the moment they are called, but they are lazy, i.e. delayed until the moment one of their results is accessed through the smart pointer. At that time, the event loop is triggered and all results are produced simultaneously.
It is therefore good practice to declare all your transformations and actions before accessing their results, allowing RDataFrame to run the loop once and produce all results in one go.
Let's say we would like to run the previous examples in parallel on several cores, dividing events fairly between cores. The only modification required to the snippets would be the addition of this line before constructing the main dataframe object:
Simple as that. More details are given below.
RDataFrame reads collections as the special type ROOT::RVec: for example, a column containing an array of floating point numbers can be read as a ROOT::RVecF. C-style arrays (with variable or static size), STL vectors and most other collection types can be read this way.
RVec is a container similar to std::vector (and can be used just like a std::vector) but it also offers a rich interface to operate on the array elements in a vectorised fashion, similarly to Python's NumPy arrays.
For example, to fill a histogram with the "pt" of selected particles for each event, Define() can be used to create a column that contains the desired array elements as follows:
And in Python:
Learn more at ROOT::VecOps::RVec.
A filter is created through a call to Filter(f, columnList) or Filter(filterString). In the first overload, f can be a function, a lambda expression, a functor class, or any other callable object. It must return a bool signalling whether the event has passed the selection (true) or not (false). It should perform "read-only" operations on the columns, and should not have side-effects (e.g. modification of an external or static variable) to ensure correctness when implicit multi-threading is active. The second overload takes a string with a valid C++ expression in which column names are used as variable names (e.g. Filter("x[0] + x[1] > 0")). This is a convenience feature that comes with a certain runtime overhead: C++ code has to be generated on the fly from this expression before using it in the event loop. See the paragraph about "Just-in-time compilation" below for more information.
RDataFrame only evaluates filters when necessary: if multiple filters are chained one after another, they are executed in order and the first one returning false causes the event to be discarded and triggers the processing of the next entry. If multiple actions or transformations depend on the same filter, that filter is not executed multiple times for each entry: after the first access it simply serves a cached result.
An optional string parameter name can be passed to the Filter() method to create a named filter. Named filters work as usual, but also keep track of how many entries they accept and reject.
Statistics are retrieved through a call to the Report() method:
Stats are stored in the same order as named filters have been added to the graph, and refer to the latest event-loop that has been run using the relevant RDataFrame.
When RDataFrame is not being used in a multi-thread environment (i.e. no call to EnableImplicitMT() was made), Range() transformations are available. These act very much like filters but instead of basing their decision on a filter expression, they rely on begin,end and stride parameters.
begin: initial entry number considered for this range.end: final entry number (excluded) considered for this range. 0 means that the range goes until the end of the dataset.stride: process one entry of the [begin, end) range every stride entries. Must be strictly greater than 0.The actual number of entries processed downstream of a Range() node will be (end - begin)/stride (or less if less entries than that are available).
Note that ranges act "locally", not based on the global entry count: Range(10,50) means "skip the first 10 entries
that reach this node*, let the next 40 entries pass, then stop processing". If a range node hangs from a filter node, and the range has a begin parameter of 10, that means the range will skip the first 10 entries that pass the preceding filter.
Ranges allow "early quitting": if all branches of execution of a functional graph reached their end value of processed entries, the event-loop is immediately interrupted. This is useful for debugging and quick data explorations.
Custom columns are created by invoking Define(name, f, columnList). As usual, f can be any callable object (function, lambda expression, functor class...); it takes the values of the columns listed in columnList (a list of strings) as parameters, in the same order as they are listed in columnList. f must return the value that will be assigned to the temporary column.
A new variable is created called name, accessible as if it was contained in the dataset from subsequent transformations/actions.
Use cases include:
An exception is thrown if the name of the new column/branch is already in use for another branch in the TTree.
It is also possible to specify the quantity to be stored in the new temporary column as a C++ expression with the method Define(name, expression). For example this invocation
will create a new column called "pt" the value of which is calculated starting from the columns px and py. The system builds a just-in-time compiled function starting from the expression after having deduced the list of necessary branches from the names of the variables specified by the user.
It is possible to create custom columns also as a function of the processing slot and entry numbers. The methods that can be invoked are:
DefineSlot(name, f, columnList). In this case the callable f has this signature R(unsigned int, T1, T2, ...): the first parameter is the slot number which ranges from 0 to ROOT::GetThreadPoolSize() - 1.DefineSlotEntry(name, f, columnList). In this case the callable f has this signature R(unsigned int, ULong64_t,
T1, T2, ...): the first parameter is the slot number while the second one the number of the entry being processed.Actions can be instant or lazy. Instant actions are executed as soon as they are called, while lazy actions are executed whenever the object they return is accessed for the first time. As a rule of thumb, actions with a return value are lazy, the others are instant.
When a lazy action is called, it returns a ROOT::RDF::RResultPtr<T>, where T is the type of the result of the action. The final result will be stored in the RResultPtr and can be retrieved by dereferencing it or via its GetValue method.
If the type of the return value of an action is a collection, e.g. std::vector<int>, you can iterate its elements directly through the wrapping RResultPtr:
An action that needs values for its computations will request it from a reader, e.g. a column created via Define or available from the input dataset. The action will request values from each column of the list of input columns (either inferred or specified by the user), in order. For example:
The Graph action is going to request first the value from column "x", then that of column "y". Specifically, the order of execution of the operations of nodes in this branch of the computation graph is guaranteed to be top to bottom.
RDataFrame applications can be executed in parallel through distributed computing frameworks on a set of remote machines thanks to the Python package ROOT.RDF.Experimental.Distributed. This experimental, Python-only package allows to scale the optimized performance RDataFrame can achieve on a single machine to multiple nodes at the same time. It is designed so that different backends can be easily plugged in, currently supporting Apache Spark and Dask. To make use of distributed RDataFrame, you only need to switch ROOT.RDataFrame with the backend-specific RDataFrame of your choice, for example:
The main goal of this package is to support running any RDataFrame application distributedly. Nonetheless, not all parts of the RDataFrame API currently work with this package. The subset that is currently available is:
with support for more operations coming in the future. Data sources other than TTree and TChain (e.g. CSV, RNTuple) are currently not supported.
In order to distribute the RDataFrame workload, you can connect to a Spark cluster you have access to through the official Spark API, then hook the connection instance to the distributed RDataFrame object like so:
If an instance of SparkContext is not provided, the default behaviour is to create one in the background for you.
Similarly, you can connect to a Dask cluster by creating your own connection object which internally operates with one of the cluster schedulers supported by Dask (more information in the Dask distributed docs):
If an instance of distributed.Client is not provided to the RDataFrame object, it will be created for you and it will run the computations in the local machine using all cores available.
A distributed RDataFrame has internal logic to define in how many chunks the input dataset will be split before sending tasks to the distributed backend. Each task reads and processes one of said chunks. The logic is backend-dependent, but generically tries to infer how many cores are available in the cluster through the connection object. The number of tasks will be equal to the inferred number of cores. There are cases where the connection object of the chosen backend doesn't have information about the actual resources of the cluster. An example of this is when using Dask to connect to a batch system. The client object created at the beginning of the application does not automatically know how many cores will be available during distributed execution, since the jobs are submitted to the batch system after the creation of the connection. In such cases, the logic is to default to process the whole dataset in 2 tasks.
The number of tasks submitted for distributed execution can be also set programmatically, by providing the optional keyword argument npartitions when creating the RDataFrame object. This parameter is accepted irrespectively of the backend used:
Note that when processing a TTree or TChain dataset, the npartitions value should not exceed the number of clusters in the dataset. The number of clusters in a TTree can be retrieved by typing rootls -lt myfile.root at a command line.
The Snapshot operation behaves slightly differently when executed distributedly. First off, it requires the path supplied to the Snapshot call to be accessible from any worker of the cluster and from the client machine (in general it should be provided as an absolute path). Another important difference is that n separate files will be produced, where n is the number of dataset partitions. As with local RDataFrame, the result of a Snapshot on a distributed RDataFrame is another distributed RDataFrame on which we can define a new computation graph and run more distributed computations.
Submitting multiple distributed RDataFrame executions is supported through the RunGraphs function. Similarly to its local counterpart, the function expects an iterable of objects representing an RDataFrame action. Each action will be triggered concurrently to send multiple computation graphs to a distributed cluster at the same time:
Every distributed backend supports this feature and graphs belonging to different backends can be still triggered with a single call to RunGraphs (e.g. it is possible to send a Spark job and a Dask job at the same time).
When calling a Histo*D operation in distributed mode, remember to pass to the function the model of the histogram to be computed, e.g. the axis range and the number of bins:
Without this, two partial histograms resulting from two distributed tasks would have incompatible binning, thus leading to errors when merging them. Failing to pass a histogram model will raise an error on the client side, before starting the distributed execution.
The live visualization feature allows real-time data representation of plots generated during the execution of a distributed RDataFrame application. It enables visualizing intermediate results as they are computed across multiple nodes of a Dask cluster by creating a canvas and continuously updating it as partial results become available.
The LiveVisualize() function can be imported from the Python package ROOT.RDF.Experimental.Distributed:
The function takes drawable objects (e.g. histograms) and optional callback functions as argument, it accepts 4 different input formats:
As pointed out before in this document, RDataFrame can transparently perform multi-threaded event loops to speed up the execution of its actions. Users have to call ROOT::EnableImplicitMT() before constructing the RDataFrame object to indicate that it should take advantage of a pool of worker threads. Each worker thread processes a distinct subset of entries, and their partial results are merged before returning the final values to the user. There are no guarantees on the order in which threads will process the batches of entries. In particular, note that this means that, for multi-thread event loops, there is no guarantee on the order in which Snapshot() will write entries: they could be scrambled with respect to the input dataset. The values of the special rdfentry_ column will also not correspond to the entry numbers in the input dataset (e.g. TChain) in multi-thread runs.
i with the number of CPUs/slots that were allocated for this job.RDataFrame operations such as Histo1D() or Snapshot() are guaranteed to work correctly in multi-thread event loops. User-defined expressions, such as strings or lambdas passed to Filter(), Define(), Foreach(), Reduce() or Aggregate() will have to be thread-safe, i.e. it should be possible to call them concurrently from different threads.
Note that simple Filter() and Define() transformations will inherently satisfy this requirement: Filter() / Define() expressions will often be pure in the functional programming sense (no side-effects, no dependency on external state), which eliminates all risks of race conditions.
In order to facilitate writing of thread-safe operations, some RDataFrame features such as Foreach(), Define() or OnPartialResult() offer thread-aware counterparts (ForeachSlot(), DefineSlot(), OnPartialResultSlot()): their only difference is that they will pass an extra slot argument (an unsigned integer) to the user-defined expression. When calling user-defined code concurrently, RDataFrame guarantees that different threads will employ different values of the slot parameter, where slot will be a number between 0 and GetNSlots() - 1. In other words, within a slot, computation runs sequentially and events are processed sequentially. Note that the same slot might be associated to different threads over the course of a single event loop, but two threads will never receive the same slot at the same time. This extra parameter might facilitate writing safe parallel code by having each thread write/modify a different processing slot, e.g. a different element of a list. See here for an example usage of ForeachSlot().
A complex analysis may require multiple separate RDataFrame computation graphs to produce all desired results. This poses the challenge that the event loops of each computation graph can be parallelized, but the different loops run sequentially, one after the other. On many-core architectures it might be desirable to run different event loops concurrently to improve resource usage. ROOT::RDF::RunGraphs() allows running multiple RDataFrame event loops concurrently:
To obtain the maximum performance out of RDataFrame, make sure to avoid just-in-time compiled versions of transformations and actions if at all possible. For instance, Filter("x > 0") requires just-in-time compilation of the corresponding C++ logic, while the equivalent Filter([](float x) { return x > 0.; }, {"x"}) does not. Similarly, Histo1D("x") requires just-in-time compilation after the type of x is retrieved from the dataset, while Histo1D<float>("x") does not; the latter spelling should be preferred for performance-critical applications.
Python applications cannot easily specify template parameters or pass C++ callables to RDataFrame. See Efficient analysis in Python for possible ways to speed up hot paths in this case.
Just-in-time compilation happens once, right before starting an event loop. To reduce the runtime cost of this step, make sure to book all operations for all RDataFrame computation graphs before the first event loop is triggered: just-in-time compilation will happen once for all code required to be generated up to that point, also across different computation graphs.
Also make sure not to count the just-in-time compilation time (which happens once before the event loop and does not depend on the size of the dataset) as part of the event loop runtime (which scales with the size of the dataset). RDataFrame has an experimental logging feature that simplifies measuring the time spent in just-in-time compilation and in the event loop (as well as providing some more interesting information). See Activating RDataFrame execution logs.
There are two reasons why RDataFrame may consume more memory than expected. Firstly, each result is duplicated for each worker thread, which e.g. in case of many (possibly multi-dimensional) histograms with fine binning can result in visible memory consumption during the event loop. The thread-local copies of the results are destroyed when the final result is produced. Reducing the number of threads or using coarser binning will reduce the memory usage.
Secondly, just-in-time compilation of string expressions or non-templated actions (see the previous paragraph) causes Cling, ROOT's C++ interpreter, to allocate some memory for the generated code that is only released at the end of the application. This commonly results in memory usage creep in long-running applications that create many RDataFrames one after the other. Possible mitigations include creating and running each RDataFrame event loop in a sub-process, or booking all operations for all different RDataFrame computation graphs before the first event loop is triggered, so that the interpreter is invoked only once for all computation graphs:
Here is a list of the most important features that have been omitted in the "Crash course" for brevity. You don't need to read all these to start using RDataFrame, but they are useful to save typing time and runtime.
Starting from ROOT v6.26, RDataFrame provides a flexible syntax to define systematic variations. This is done in two steps: a) variations for one or more existing columns are registered via Vary() and b) variations of normal RDataFrame results are extracted with a call to VariationsFor(). In between these steps, no other change to the analysis code is required: the presence of systematic variations for certain columns is automatically propagated through filters, defines and actions, and RDataFrame will take these dependencies into account when producing varied results. VariationsFor() is included in header ROOT/RDFHelpers.hxx, which compiled C++ programs must include explicitly.
An example usage of Vary() and VariationsFor() in C++:
A list of variation "tags" is passed as last argument to Vary(): they give a name to the varied values that are returned as elements of an RVec of the appropriate type. The number of variation tags must correspond to the number of elements the RVec returned by the expression (2 in the example above: the first element will correspond to tag "down", the second to tag "up"). The full variation name will be composed of the varied column name and the variation tags (e.g. "pt:down", "pt:up" in this example). Python usage looks similar.
Note how we use the "pt" column as usual in the Filter() and Define() calls and we simply use "x" as the value to fill the resulting histogram. To produce the varied results, RDataFrame will automatically execute the Filter and Define calls for each variation and fill the histogram with values and cuts that depend on the variation.
There is no limitation to the complexity of a Vary() expression, and just like for Define() and Filter() calls users are not limited to string expressions but they can also pass any valid C++ callable, including lambda functions and complex functors. The callable can be applied to zero or more existing columns and it will always receive their nominal values in input.
In the following Python snippet we use the Vary() signature that allows varying multiple columns simultaneously or "in lockstep":
The expression returns an RVec of two RVecs: each inner vector contains the varied values for one column, and the inner vectors follow the same ordering as the column names passed as first argument. Besides the variation tags, in this case we also have to explicitly pass a variation name as there is no one column name that can be used as default.
The call above will produce variations "ptAndEta:down" and "ptAndEta:up".
Even if a result depends on multiple variations, only one is applied at a time, i.e. there will be no result produced by applying multiple systematic variations at the same time. For example, in the following example snippet, the RResultMap instance all_h will contain keys "nominal", "pt:down", "pt:up", "eta:0", "eta:1", but no "pt:up&&eta:0" or similar:
Note how we passed the integer 2 instead of a list of variation tags to the second Vary() invocation: this is a shorthand that automatically generates tags 0 to N-1 (in this case 0 and 1).
ROOT::RDF::Experimental namespace, to indicate that these interfaces might still evolve and improve based on user feedback. We expect that some aspects of the related programming model will be streamlined in future versions.RDataFrame variables/nodes are relatively cheap to copy and it's possible to both pass them to (or move them into) functions and to return them from functions. However, in general each dataframe node will have a different C++ type, which includes all available compile-time information about what that node does. One way to cope with this complication is to use template functions and/or C++14 auto return types:
A possibly simpler, C++11-compatible alternative is to take advantage of the fact that any dataframe node can be converted (implicitly or via an explicit cast) to the common type ROOT::RDF::RNode:
The conversion to ROOT::RDF::RNode is cheap, but it will introduce an extra virtual call during the RDataFrame event loop (in most cases, the resulting performance impact should be negligible). Python users can perform the conversion with the helper function ROOT.RDF.AsRNode.
ROOT::RDF::RNode also makes it simple to store RDataFrame nodes in collections, e.g. a std::vector<RNode> or a std::map<std::string, RNode>:
It's possible to schedule execution of arbitrary functions (callbacks) during the event loop. Callbacks can be used e.g. to inspect partial results of the analysis while the event loop is running, drawing a partially-filled histogram every time a certain number of new entries is processed, or displaying a progress bar while the event loop runs.
For example one can draw an up-to-date version of a result histogram every 100 entries like this:
Callbacks are registered to a ROOT::RDF::RResultPtr and must be callables that takes a reference to the result type as argument and return nothing. RDataFrame will invoke registered callbacks passing partial action results as arguments to them (e.g. a histogram filled with a part of the selected events).
Read more on ROOT::RDF::RResultPtr::OnPartialResult() and ROOT::RDF::RResultPtr::OnPartialResultSlot().
When constructing an RDataFrame object, it is possible to specify a default column list for your analysis, in the usual form of a list of strings representing branch/column names. The default column list will be used as a fallback whenever a list specific to the transformation/action is not present. RDataFrame will take as many of these columns as needed, ignoring trailing extra names if present.
Every instance of RDataFrame is created with two special columns called rdfentry_ and rdfslot_. The rdfentry_ column is of type ULong64_t and it holds the current entry number while rdfslot_ is an unsigned int holding the index of the current data processing slot. For backwards compatibility reasons, the names tdfentry_ and tdfslot_ are also accepted. These columns are ignored by operations such as Cache or Snapshot.
rdfentry_ do not correspond to what would be the entry numbers of a TChain constructed over the same set of ROOT files, as the entries are processed in an unspecified order.C++ is a statically typed language: all types must be known at compile-time. This includes the types of the TTree branches we want to work on. For filters, defined columns and some of the actions, column types are deduced from the signature of the relevant filter function/temporary column expression/action function:
If we specify an incorrect type for one of the columns, an exception with an informative message will be thrown at runtime, when the column value is actually read from the dataset: RDataFrame detects type mismatches. The same would happen if we swapped the order of "b1" and "b2" in the column list passed to Filter().
Certain actions, on the other hand, do not take a function as argument (e.g. Histo1D()), so we cannot deduce the type of the column at compile-time. In this case RDataFrame infers the type of the column from the TTree itself. This is why we never needed to specify the column types for all actions in the above snippets.
When the column type is not a common one such as int, double, char or float it is nonetheless good practice to specify it as a template parameter to the action itself, like this:
Deducing types at runtime requires the just-in-time compilation of the relevant actions, which has a small runtime overhead, so specifying the type of the columns as template parameters to the action is good practice when performance is a goal.
When strings are passed as expressions to Filter() or Define(), fundamental types are passed as constants. This avoids certaincommon mistakes such as typing x = 0 rather than x == 0:
RDataFrame strives to offer a comprehensive set of standard actions that can be performed on each event. At the same time, it allows users to inject their own action code to perform arbitrarily complex data reductions.
Through the Book() method, users can implement a custom action and have access to the same features that built-in RDataFrame actions have, e.g. hooks to events related to the start, end and execution of the event loop, or the possibility to return a lazy RResultPtr to an arbitrary type of result:
See the Book() method for more information and this tutorial for a more complete example.
Foreach() takes a callable (lambda expression, free function, functor...) and a list of columns and executes the callable on the values of those columns for each event that passes all upstream selections. It can be used to perform actions that are not already available in the interface. For example, the following snippet evaluates the root mean square of column "x":
In multi-thread runs, users are responsible for the thread-safety of the expression passed to Foreach(): thread will execute the expression concurrently. The code above would need to employ some resource protection mechanism to ensure non-concurrent writing of rms; but this is probably too much head-scratch for such a simple operation.
ForeachSlot() can help in this situation. It is an alternative version of Foreach() for which the function takes an additional "processing slot" parameter besides the columns it should be applied to. RDataFrame guarantees that ForeachSlot() will invoke the user expression with different slot parameters for different concurrent executions (see Special helper columns: rdfentry_ and rdfslot_ for more information on the slot parameter). We can take advantage of ForeachSlot() to evaluate a thread-safe root mean square of column "x":
Notice how we created one double variable for each processing slot and later merged their results via std::accumulate.
Vertically concatenating multiple trees that have the same columns (creating a logical dataset with the same columns and more rows) is trivial in RDataFrame: just pass the tree name and a list of file names to RDataFrame's constructor, or create a TChain out of the desired trees and pass that to RDataFrame.
Horizontal concatenations of trees or chains (creating a logical dataset with the same number of rows and the union of the columns of multiple trees) leverages TTree's "friend" mechanism.
Simple joins of trees that do not have the same number of rows are also possible with indexed friend trees (see below).
To use friend trees in RDataFrame, set up trees with the appropriate relationships and then instantiate an RDataFrame with the main tree:
The same applies for TChains. Columns coming from the friend trees can be referred to by their full name, like in the example above, or the friend tree name can be omitted in case the column name is not ambiguous (e.g. "MyCol" could be used instead of "myFriend.MyCol" in the example above if there is no column "MyCol" in the main tree).
Indexed friend trees provide a way to perform simple joins of multiple trees over a common column. When a certain entry in the main tree (or chain) is loaded, the friend trees (or chains) will then load an entry where the "index" columns have a value identical to the one in the main one. For example, in Python:
RDataFrame supports indexed friend TTrees from ROOT v6.24 in single-thread mode and from v6.28/02 in multi-thread mode.
RDataFrame can be interfaced with RDataSources. The ROOT::RDF::RDataSource interface defines an API that RDataFrame can use to read arbitrary columnar data formats.
RDataFrame calls into concrete RDataSource implementations to retrieve information about the data, retrieve (thread-local) readers or "cursors" for selected columns and to advance the readers to the desired data entry. Some predefined RDataSources are natively provided by ROOT such as the ROOT::RDF::RCsvDS which allows to read comma separated files:
See also FromNumpy (Python-only), FromRNTuple(), FromArrow(), FromSqlite().
As we saw, transformed dataframes can be stored as variables and reused multiple times to create modified versions of the dataset. This implicitly defines a computation graph in which several paths of filtering/creation of columns are executed simultaneously, and finally aggregated results are produced.
RDataFrame detects when several actions use the same filter or the same defined column, and only evaluates each filter or defined column once per event, regardless of how many times that result is used down the computation graph. Objects read from each column are built once and never copied, for maximum efficiency. When "upstream" filters are not passed, subsequent filters, temporary column expressions and actions are not evaluated, so it might be advisable to put the strictest filters first in the graph.
It is possible to print the computation graph from any node to obtain a DOT (graphviz) representation either on the standard output or in a file.
Invoking the function ROOT::RDF::SaveGraph() on any node that is not the head node, the computation graph of the branch the node belongs to is printed. By using the head node, the entire computation graph is printed.
Following there is an example of usage:
The generated graph can be rendered using one of the graphviz filters, e.g. dot. For instance, the image below can be generated with the following command:

RDataFrame has experimental support for verbose logging of the event loop runtimes and other interesting related information. It is activated as follows:
or in Python:
More information (e.g. start and end of each multi-thread task) is printed using ELogLevel.kDebug and even more (e.g. a full dump of the generated code that RDataFrame just-in-time-compiles) using ELogLevel.kDebug+10.
RDataFrame can be created using a dataset specification JSON file:
The input dataset specification JSON file needs to be provided by the user and it describes all necessary samples and their associated metadata information. The main required key is the "samples" (at least one sample is needed) and the required sub-keys for each sample are "trees" and "files". Additionally, one can specify a metadata dictionary for each sample in the "metadata" key.
A simple example for the formatting of the specification in the JSON file is the following:
The metadata information from the specification file can be then accessed using the DefinePerSample function. For example, to access luminosity information (stored as a double):
or sample_category information (stored as a string):
or directly the filename:
An example implementation of the "FromSpec" method is available in tutorial: df106_HiggstoFourLeptons.py, which also provides a corresponding exemplary JSON file for the dataset specification.
A progress bar showing the processed event statistics can be added to any RDataFrame program. The event statistics include elapsed time, currently processed file, currently processed events, the rate of event processing and an estimated remaining time (per file being processed). It is recorded and printed in the terminal every m events and every n seconds (by default m = 1000 and n = 1). The ProgressBar can be also added when the multithread (MT) mode is enabled.
ProgressBar is added after creating the dataframe object (df):
Alternatively, RDataFrame can be cast to an RNode first, giving the user more flexibility For example, it can be called at any computational node, such as Filter or Define, not only the head node, with no change to the ProgressBar function itself:
Examples of implemented progress bars can be seen by running Higgs to Four Lepton tutorial and Dimuon tutorial.
You can use RDataFrame in Python thanks to the dynamic Python/C++ translation of PyROOT. In general, the interface is the same as for C++, a simple example follows.
In the simple example that was shown above, a C++ expression is passed to the Filter() operation as a string ("x > 0"), even if we call the method from Python. Indeed, under the hood, the analysis computations run in C++, while Python is just the interface language.
To perform more complex operations that don't fit into a simple expression string, you can just-in-time compile C++ functions - via the C++ interpreter cling - and use those functions in an expression. See the following snippet for an example:
To increase the performance even further, you can also pre-compile a C++ library with full code optimizations and load the function into the RDataFrame computation as follows.
A more thorough explanation of how to use C++ code from Python can be found in the PyROOT manual.
ROOT also offers the option to compile Python functions with fundamental types and arrays thereof using Numba. Such compiled functions can then be used in a C++ expression provided to RDataFrame.
The function to be compiled should be decorated with ROOT.Numba.Declare, which allows to specify the parameter and return types. See the following snippet for a simple example or the full tutorial here.
It also works with collections: RVec objects of fundamental types can be transparently converted to/from numpy arrays:
Note that this functionality requires the Python packages numba and cffi to be installed.
Eventually, you probably would like to inspect the content of the RDataFrame or process the data further with Python libraries. For this purpose, we provide the AsNumpy() function, which returns the columns of your RDataFrame as a dictionary of NumPy arrays. See a simple example below or a full tutorial here.
In case you have data in NumPy arrays in Python and you want to process the data with ROOT, you can easily create an RDataFrame using ROOT.RDF.FromNumpy. The factory function accepts a dictionary where the keys are the column names and the values are NumPy arrays, and returns a new RDataFrame with the provided columns.
Only arrays of fundamental types (integers and floating point values) are supported and the arrays must have the same length. Data is read directly from the arrays: no copies are performed.
The Histo1D(), Histo2D(), Histo3D(), Profile1D() and Profile2D() methods return histograms and profiles, respectively, which can be constructed using a model argument.
In Python, we can specify the arguments for the constructor of such histogram or profile model with a Python tuple, as shown in the example below:
The ROOT::RDF::AsRNode function casts an RDataFrame node to the generic ROOT::RDF::RNode type. From Python, it can be used to pass any RDataFrame node as an argument of a C++ function, as shown below:
Definition at line 41 of file RDataFrame.hxx.
Public Types | |
| using | ColumnNames_t = ROOT::RDF::ColumnNames_t |
Public Member Functions | |
| RDataFrame (ROOT::RDF::Experimental::RDatasetSpec spec) | |
| Build dataframe from an RDatasetSpec object. | |
| RDataFrame (std::string_view treeName, ::TDirectory *dirPtr, const ColumnNames_t &defaultColumns={}) | |
| RDataFrame (std::string_view treename, const std::vector< std::string > &filenames, const ColumnNames_t &defaultColumns={}) | |
| Build the dataframe. | |
| RDataFrame (std::string_view treename, std::initializer_list< std::string > filenames, const ColumnNames_t &defaultColumns={}) | |
| RDataFrame (std::string_view treeName, std::string_view filenameglob, const ColumnNames_t &defaultColumns={}) | |
| Build the dataframe. | |
| RDataFrame (std::unique_ptr< ROOT::RDF::RDataSource >, const ColumnNames_t &defaultColumns={}) | |
| Build dataframe associated to data source. | |
| RDataFrame (TTree &tree, const ColumnNames_t &defaultColumns={}) | |
| Build the dataframe. | |
| RDataFrame (ULong64_t numEntries) | |
| Build a dataframe that generates numEntries entries. | |
 Public Member Functions inherited from ROOT::RDF::RInterface< RDFDetail::RLoopManager > Public Member Functions inherited from ROOT::RDF::RInterface< RDFDetail::RLoopManager > | |
| RInterface (const RInterface &)=default | |
| Copy-ctor for RInterface. | |
| RInterface (const std::shared_ptr< RLoopManager > &proxied) | |
| Build a RInterface from a RLoopManager. | |
| RInterface (RInterface &&)=default | |
| Move-ctor for RInterface. | |
| RResultPtr< U > | Aggregate (AccFun aggregator, MergeFun merger, std::string_view columnName, const U &aggIdentity) |
| Execute a user-defined accumulation operation on the processed column values in each processing slot. | |
| RResultPtr< U > | Aggregate (AccFun aggregator, MergeFun merger, std::string_view columnName="") |
| Execute a user-defined accumulation operation on the processed column values in each processing slot. | |
| RInterface< RDFDetail::RLoopManager, DS_t > | Alias (std::string_view alias, std::string_view columnName) |
| Allow to refer to a column with a different name. | |
| RResultPtr< typename std::decay_t< Helper >::Result_t > | Book (Helper &&helper, const ColumnNames_t &columns={}) |
| Book execution of a custom action using a user-defined helper object. | |
| RInterface< RLoopManager > | Cache (const ColumnNames_t &columnList) |
| Save selected columns in memory. | |
| RInterface< RLoopManager > | Cache (const ColumnNames_t &columnList) |
| Save selected columns in memory. | |
| RInterface< RLoopManager > | Cache (std::initializer_list< std::string > columnList) |
| Save selected columns in memory. | |
| RInterface< RLoopManager > | Cache (std::string_view columnNameRegexp="") |
| Save selected columns in memory. | |
| RResultPtr< ULong64_t > | Count () |
| Return the number of entries processed (lazy action). | |
| RInterface< RDFDetail::RLoopManager, DS_t > | Define (std::string_view name, F expression, const ColumnNames_t &columns={}) |
| Define a new column. | |
| RInterface< RDFDetail::RLoopManager, DS_t > | Define (std::string_view name, std::string_view expression) |
| Define a new column. | |
| RInterface< RDFDetail::RLoopManager, DS_t > | DefinePerSample (std::string_view name, F expression) |
| Define a new column that is updated when the input sample changes. | |
| RInterface< RDFDetail::RLoopManager, DS_t > | DefinePerSample (std::string_view name, std::string_view expression) |
| Define a new column that is updated when the input sample changes. | |
| RInterface< RDFDetail::RLoopManager, DS_t > | DefineSlot (std::string_view name, F expression, const ColumnNames_t &columns={}) |
| Define a new column with a value dependent on the processing slot. | |
| RInterface< RDFDetail::RLoopManager, DS_t > | DefineSlotEntry (std::string_view name, F expression, const ColumnNames_t &columns={}) |
| Define a new column with a value dependent on the processing slot and the current entry. | |
| RResultPtr< RDisplay > | Display (const ColumnNames_t &columnList, size_t nRows=5, size_t nMaxCollectionElements=10) |
| Provides a representation of the columns in the dataset. | |
| RResultPtr< RDisplay > | Display (const ColumnNames_t &columnList, size_t nRows=5, size_t nMaxCollectionElements=10) |
| Provides a representation of the columns in the dataset. | |
| RResultPtr< RDisplay > | Display (std::initializer_list< std::string > columnList, size_t nRows=5, size_t nMaxCollectionElements=10) |
| Provides a representation of the columns in the dataset. | |
| RResultPtr< RDisplay > | Display (std::string_view columnNameRegexp="", size_t nRows=5, size_t nMaxCollectionElements=10) |
| Provides a representation of the columns in the dataset. | |
| RResultPtr< std::decay_t< T > > | Fill (T &&model, const ColumnNames_t &columnList) |
Return an object of type T on which T::Fill will be called once per event (lazy action). | |
| RInterface< RDFDetail::RFilter< F, RDFDetail::RLoopManager >, DS_t > | Filter (F f, const ColumnNames_t &columns={}, std::string_view name="") |
| Append a filter to the call graph. | |
| RInterface< RDFDetail::RFilter< F, RDFDetail::RLoopManager >, DS_t > | Filter (F f, const std::initializer_list< std::string > &columns) |
| Append a filter to the call graph. | |
| RInterface< RDFDetail::RFilter< F, RDFDetail::RLoopManager >, DS_t > | Filter (F f, std::string_view name) |
| Append a filter to the call graph. | |
| RInterface< RDFDetail::RJittedFilter, DS_t > | Filter (std::string_view expression, std::string_view name="") |
| Append a filter to the call graph. | |
| void | Foreach (F f, const ColumnNames_t &columns={}) |
| Execute a user-defined function on each entry (instant action). | |
| void | ForeachSlot (F f, const ColumnNames_t &columns={}) |
| Execute a user-defined function requiring a processing slot index on each entry (instant action). | |
| std::vector< std::string > | GetFilterNames () |
| Returns the names of the filters created. | |
| RResultPtr<::TGraph > | Graph (std::string_view x="", std::string_view y="") |
| Fill and return a TGraph object (lazy action). | |
| RResultPtr<::TGraphAsymmErrors > | GraphAsymmErrors (std::string_view x="", std::string_view y="", std::string_view exl="", std::string_view exh="", std::string_view eyl="", std::string_view eyh="") |
| Fill and return a TGraphAsymmErrors object (lazy action). | |
| RResultPtr<::TH1D > | Histo1D (const TH1DModel &model, std::string_view vName, std::string_view wName) |
| Fill and return a one-dimensional histogram with the weighted values of a column (lazy action). | |
| RResultPtr<::TH1D > | Histo1D (const TH1DModel &model={"", "", 128u, 0., 0.}) |
| Fill and return a one-dimensional histogram with the weighted values of a column (lazy action). | |
| RResultPtr<::TH1D > | Histo1D (const TH1DModel &model={"", "", 128u, 0., 0.}, std::string_view vName="") |
| Fill and return a one-dimensional histogram with the values of a column (lazy action). | |
| RResultPtr<::TH1D > | Histo1D (std::string_view vName) |
| Fill and return a one-dimensional histogram with the values of a column (lazy action). | |
| RResultPtr<::TH1D > | Histo1D (std::string_view vName, std::string_view wName) |
| Fill and return a one-dimensional histogram with the weighted values of a column (lazy action). | |
| RResultPtr<::TH2D > | Histo2D (const TH2DModel &model) |
| RResultPtr<::TH2D > | Histo2D (const TH2DModel &model, std::string_view v1Name, std::string_view v2Name, std::string_view wName) |
| Fill and return a weighted two-dimensional histogram (lazy action). | |
| RResultPtr<::TH2D > | Histo2D (const TH2DModel &model, std::string_view v1Name="", std::string_view v2Name="") |
| Fill and return a two-dimensional histogram (lazy action). | |
| RResultPtr<::TH3D > | Histo3D (const TH3DModel &model) |
| RResultPtr<::TH3D > | Histo3D (const TH3DModel &model, std::string_view v1Name, std::string_view v2Name, std::string_view v3Name, std::string_view wName) |
| Fill and return a three-dimensional histogram (lazy action). | |
| RResultPtr<::TH3D > | Histo3D (const TH3DModel &model, std::string_view v1Name="", std::string_view v2Name="", std::string_view v3Name="") |
| Fill and return a three-dimensional histogram (lazy action). | |
| RResultPtr<::THnD > | HistoND (const THnDModel &model, const ColumnNames_t &columnList) |
| Fill and return an N-dimensional histogram (lazy action). | |
| RResultPtr<::THnD > | HistoND (const THnDModel &model, const ColumnNames_t &columnList) |
| Fill and return an N-dimensional histogram (lazy action). | |
| RResultPtr< RDFDetail::MaxReturnType_t< T > > | Max (std::string_view columnName="") |
| Return the maximum of processed column values (lazy action). | |
| RResultPtr< double > | Mean (std::string_view columnName="") |
| Return the mean of processed column values (lazy action). | |
| RResultPtr< RDFDetail::MinReturnType_t< T > > | Min (std::string_view columnName="") |
| Return the minimum of processed column values (lazy action). | |
| operator RNode () const | |
| Cast any RDataFrame node to a common type ROOT::RDF::RNode. | |
| RInterface & | operator= (const RInterface &)=default |
| Copy-assignment operator for RInterface. | |
| RInterface & | operator= (RInterface &&)=default |
| Move-assignment operator for RInterface. | |
| RResultPtr<::TProfile > | Profile1D (const TProfile1DModel &model) |
| Fill and return a one-dimensional profile (lazy action). | |
| RResultPtr<::TProfile > | Profile1D (const TProfile1DModel &model, std::string_view v1Name, std::string_view v2Name, std::string_view wName) |
| Fill and return a one-dimensional profile (lazy action). | |
| RResultPtr<::TProfile > | Profile1D (const TProfile1DModel &model, std::string_view v1Name="", std::string_view v2Name="") |
| Fill and return a one-dimensional profile (lazy action). | |
| RResultPtr<::TProfile2D > | Profile2D (const TProfile2DModel &model) |
| Fill and return a two-dimensional profile (lazy action). | |
| RResultPtr<::TProfile2D > | Profile2D (const TProfile2DModel &model, std::string_view v1Name, std::string_view v2Name, std::string_view v3Name, std::string_view wName) |
| Fill and return a two-dimensional profile (lazy action). | |
| RResultPtr<::TProfile2D > | Profile2D (const TProfile2DModel &model, std::string_view v1Name="", std::string_view v2Name="", std::string_view v3Name="") |
| Fill and return a two-dimensional profile (lazy action). | |
| RInterface< RDFDetail::RRange< RDFDetail::RLoopManager >, DS_t > | Range (unsigned int begin, unsigned int end, unsigned int stride=1) |
| Creates a node that filters entries based on range: [begin, end). | |
| RInterface< RDFDetail::RRange< RDFDetail::RLoopManager >, DS_t > | Range (unsigned int end) |
| Creates a node that filters entries based on range. | |
| RInterface< RDFDetail::RLoopManager, DS_t > | Redefine (std::string_view name, F expression, const ColumnNames_t &columns={}) |
| Overwrite the value and/or type of an existing column. | |
| RInterface< RDFDetail::RLoopManager, DS_t > | Redefine (std::string_view name, std::string_view expression) |
| Overwrite the value and/or type of an existing column. | |
| RInterface< RDFDetail::RLoopManager, DS_t > | RedefineSlot (std::string_view name, F expression, const ColumnNames_t &columns={}) |
| Overwrite the value and/or type of an existing column. | |
| RInterface< RDFDetail::RLoopManager, DS_t > | RedefineSlotEntry (std::string_view name, F expression, const ColumnNames_t &columns={}) |
| Overwrite the value and/or type of an existing column. | |
| RResultPtr< T > | Reduce (F f, std::string_view columnName, const T &redIdentity) |
| Execute a user-defined reduce operation on the values of a column. | |
| RResultPtr< T > | Reduce (F f, std::string_view columnName="") |
| Execute a user-defined reduce operation on the values of a column. | |
| RResultPtr< RCutFlowReport > | Report () |
| Gather filtering statistics. | |
| RResultPtr< RInterface< RLoopManager > > | Snapshot (std::string_view treename, std::string_view filename, const ColumnNames_t &columnList, const RSnapshotOptions &options=RSnapshotOptions()) |
Save selected columns to disk, in a new TTree treename in file filename. | |
| RResultPtr< RInterface< RLoopManager > > | Snapshot (std::string_view treename, std::string_view filename, const ColumnNames_t &columnList, const RSnapshotOptions &options=RSnapshotOptions()) |
Save selected columns to disk, in a new TTree treename in file filename. | |
| RResultPtr< RInterface< RLoopManager > > | Snapshot (std::string_view treename, std::string_view filename, std::initializer_list< std::string > columnList, const RSnapshotOptions &options=RSnapshotOptions()) |
Save selected columns to disk, in a new TTree treename in file filename. | |
| RResultPtr< RInterface< RLoopManager > > | Snapshot (std::string_view treename, std::string_view filename, std::string_view columnNameRegexp="", const RSnapshotOptions &options=RSnapshotOptions()) |
Save selected columns to disk, in a new TTree treename in file filename. | |
| RResultPtr< TStatistic > | Stats (std::string_view value, std::string_view weight) |
| Return a TStatistic object, filled once per event (lazy action). | |
| RResultPtr< TStatistic > | Stats (std::string_view value="") |
| Return a TStatistic object, filled once per event (lazy action). | |
| RResultPtr< double > | StdDev (std::string_view columnName="") |
| Return the unbiased standard deviation of processed column values (lazy action). | |
| RResultPtr< RDFDetail::SumReturnType_t< T > > | Sum (std::string_view columnName="", const RDFDetail::SumReturnType_t< T > &initValue=RDFDetail::SumReturnType_t< T >{}) |
| Return the sum of processed column values (lazy action). | |
| RResultPtr< COLL > | Take (std::string_view column="") |
| Return a collection of values of a column (lazy action, returns a std::vector by default). | |
| RInterface< RDFDetail::RLoopManager, DS_t > | Vary (const std::vector< std::string > &colNames, F &&expression, const ColumnNames_t &inputColumns, const std::vector< std::string > &variationTags, std::string_view variationName) |
| Register a systematic variation that affects multiple columns simultaneously. | |
| RInterface< RDFDetail::RLoopManager, DS_t > | Vary (const std::vector< std::string > &colNames, F &&expression, const ColumnNames_t &inputColumns, std::size_t nVariations, std::string_view variationName) |
| Register systematic variations for one or more existing columns using auto-generated tags. | |
| RInterface< RDFDetail::RLoopManager, DS_t > | Vary (const std::vector< std::string > &colNames, std::string_view expression, const std::vector< std::string > &variationTags, std::string_view variationName) |
| Register systematic variations for one or more existing columns. | |
| RInterface< RDFDetail::RLoopManager, DS_t > | Vary (const std::vector< std::string > &colNames, std::string_view expression, std::size_t nVariations, std::string_view variationName) |
| Register systematic variations for one or more existing columns. | |
| RInterface< RDFDetail::RLoopManager, DS_t > | Vary (std::initializer_list< std::string > colNames, F &&expression, const ColumnNames_t &inputColumns, const std::vector< std::string > &variationTags, std::string_view variationName) |
| Overload to avoid ambiguity between C++20 string, vector<string> construction from init list. | |
| RInterface< RDFDetail::RLoopManager, DS_t > | Vary (std::initializer_list< std::string > colNames, F &&expression, const ColumnNames_t &inputColumns, std::size_t nVariations, std::string_view variationName) |
| Overload to avoid ambiguity between C++20 string, vector<string> construction from init list. | |
| RInterface< RDFDetail::RLoopManager, DS_t > | Vary (std::initializer_list< std::string > colNames, std::string_view expression, std::size_t nVariations, std::string_view variationName) |
| Overload to avoid ambiguity between C++20 string, vector<string> construction from init list. | |
| RInterface< RDFDetail::RLoopManager, DS_t > | Vary (std::string_view colName, F &&expression, const ColumnNames_t &inputColumns, const std::vector< std::string > &variationTags, std::string_view variationName="") |
| Register systematic variations for an existing column. | |
| RInterface< RDFDetail::RLoopManager, DS_t > | Vary (std::string_view colName, F &&expression, const ColumnNames_t &inputColumns, std::size_t nVariations, std::string_view variationName="") |
| Register systematic variations for an existing columns using auto-generated variation tags. | |
| RInterface< RDFDetail::RLoopManager, DS_t > | Vary (std::string_view colName, std::string_view expression, const std::vector< std::string > &variationTags, std::string_view variationName="") |
| Register systematic variations for an existing column. | |
| RInterface< RDFDetail::RLoopManager, DS_t > | Vary (std::string_view colName, std::string_view expression, std::size_t nVariations, std::string_view variationName="") |
| Register systematic variations for an existing column. | |
 Public Member Functions inherited from ROOT::RDF::RInterfaceBase Public Member Functions inherited from ROOT::RDF::RInterfaceBase | |
| RInterfaceBase (RDFDetail::RLoopManager &lm, const RDFInternal::RColumnRegister &colRegister) | |
| RInterfaceBase (std::shared_ptr< RDFDetail::RLoopManager > lm) | |
| RDFDescription | Describe () |
| Return information about the dataframe. | |
| ColumnNames_t | GetColumnNames () |
| Returns the names of the available columns. | |
| std::string | GetColumnType (std::string_view column) |
| Return the type of a given column as a string. | |
| ColumnNames_t | GetDefinedColumnNames () |
| Returns the names of the defined columns. | |
| unsigned int | GetNFiles () |
| unsigned int | GetNRuns () const |
| Gets the number of event loops run. | |
| unsigned int | GetNSlots () const |
| Gets the number of data processing slots. | |
| RVariationsDescription | GetVariations () const |
| Return a descriptor for the systematic variations registered in this branch of the computation graph. | |
| bool | HasColumn (std::string_view columnName) |
| Checks if a column is present in the dataset. | |
Additional Inherited Members | |
 Protected Member Functions inherited from ROOT::RDF::RInterface< RDFDetail::RLoopManager > Protected Member Functions inherited from ROOT::RDF::RInterface< RDFDetail::RLoopManager > | |
| RInterface (const std::shared_ptr< RDFDetail::RLoopManager > &proxied, RLoopManager &lm, const RDFInternal::RColumnRegister &colRegister) | |
| const std::shared_ptr< RDFDetail::RLoopManager > & | GetProxiedPtr () const |
 Protected Member Functions inherited from ROOT::RDF::RInterfaceBase Protected Member Functions inherited from ROOT::RDF::RInterfaceBase | |
| void | AddDefaultColumns () |
| template<typename... ColumnTypes> | |
| void | CheckAndFillDSColumns (ColumnNames_t validCols, TTraits::TypeList< ColumnTypes... > typeList) |
| void | CheckIMTDisabled (std::string_view callerName) |
| template<typename ActionTag , typename... ColTypes, typename ActionResultType , typename RDFNode , typename HelperArgType = ActionResultType, std::enable_if_t< RDFInternal::RNeedJitting< ColTypes... >::value, int > = 0> | |
| RResultPtr< ActionResultType > | CreateAction (const ColumnNames_t &columns, const std::shared_ptr< ActionResultType > &r, const std::shared_ptr< HelperArgType > &helperArg, const std::shared_ptr< RDFNode > &proxiedPtr, const int nColumns=-1) |
| Create RAction object, return RResultPtr for the action Overload for the case in which one or more column types were not specified (RTTI + jitting). | |
| template<typename ActionTag , typename... ColTypes, typename ActionResultType , typename RDFNode , typename HelperArgType = ActionResultType, std::enable_if_t<!RDFInternal::RNeedJitting< ColTypes... >::value, int > = 0> | |
| RResultPtr< ActionResultType > | CreateAction (const ColumnNames_t &columns, const std::shared_ptr< ActionResultType > &r, const std::shared_ptr< HelperArgType > &helperArg, const std::shared_ptr< RDFNode > &proxiedPtr, const int=-1) |
| Create RAction object, return RResultPtr for the action Overload for the case in which all column types were specified (no jitting). | |
| std::string | DescribeDataset () const |
| ColumnNames_t | GetColumnTypeNamesList (const ColumnNames_t &columnList) |
| RDFDetail::RLoopManager * | GetLoopManager () const |
| ColumnNames_t | GetValidatedColumnNames (const unsigned int nColumns, const ColumnNames_t &columns) |
| template<typename RetType > | |
| void | SanityChecksForVary (const std::vector< std::string > &colNames, const std::vector< std::string > &variationTags, std::string_view variationName) |
 Protected Attributes inherited from ROOT::RDF::RInterfaceBase Protected Attributes inherited from ROOT::RDF::RInterfaceBase | |
| RDFInternal::RColumnRegister | fColRegister |
| Contains the columns defined up to this node. | |
| RDataSource * | fDataSource = nullptr |
| Non-owning pointer to a data-source object. Null if no data-source. RLoopManager has ownership of the object. | |
| RDFDetail::RLoopManager * | fLoopManager |
| < The RLoopManager at the root of this computation graph. Never null. | |
#include <ROOT/RDataFrame.hxx>
Definition at line 43 of file RDataFrame.hxx.
| ROOT::RDataFrame::RDataFrame | ( | std::string_view | treeName, |
| std::string_view | filenameglob, | ||
| const ColumnNames_t & | defaultColumns = {} |
||
| ) |
Build the dataframe.
| [in] | treeName | Name of the tree contained in the directory |
| [in] | filenameglob | TDirectory where the tree is stored, e.g. a TFile. |
| [in] | defaultColumns | Collection of default columns. |
The filename glob supports the same type of expressions as TChain::Add(), and it is passed as-is to TChain's constructor.
The default columns are looked at in case no column is specified in the booking of actions or transformations.
Definition at line 1581 of file RDataFrame.cxx.
| ROOT::RDataFrame::RDataFrame | ( | std::string_view | treeName, |
| const std::vector< std::string > & | fileglobs, | ||
| const ColumnNames_t & | defaultColumns = {} |
||
| ) |
Build the dataframe.
| [in] | treeName | Name of the tree contained in the directory |
| [in] | fileglobs | Collection of file names of filename globs |
| [in] | defaultColumns | Collection of default columns. |
The filename globs support the same type of expressions as TChain::Add(), and each glob is passed as-is to TChain's constructor.
The default columns are looked at in case no column is specified in the booking of actions or transformations.
Definition at line 1602 of file RDataFrame.cxx.
|
inline |
Definition at line 47 of file RDataFrame.hxx.
| ROOT::RDataFrame::RDataFrame | ( | std::string_view | treeName, |
| ::TDirectory * | dirPtr, | ||
| const ColumnNames_t & | defaultColumns = {} |
||
| ) |
| ROOT::RDataFrame::RDataFrame | ( | TTree & | tree, |
| const ColumnNames_t & | defaultColumns = {} |
||
| ) |
Build the dataframe.
| [in] | tree | The tree or chain to be studied. |
| [in] | defaultColumns | Collection of default column names to fall back to when none is specified. |
The default columns are looked at in case no column is specified in the booking of actions or transformations.
Definition at line 1621 of file RDataFrame.cxx.
| ROOT::RDataFrame::RDataFrame | ( | ULong64_t | numEntries | ) |
Build a dataframe that generates numEntries entries.
| [in] | numEntries | The number of entries to generate. |
An empty-source dataframe constructed with a number of entries will generate those entries on the fly when some action is triggered, and it will do so for all the previously-defined columns.
Definition at line 1634 of file RDataFrame.cxx.
| ROOT::RDataFrame::RDataFrame | ( | std::unique_ptr< ROOT::RDF::RDataSource > | ds, |
| const ColumnNames_t & | defaultColumns = {} |
||
| ) |
Build dataframe associated to data source.
| [in] | ds | The data source object. |
| [in] | defaultColumns | Collection of default column names to fall back to when none is specified. |
A dataframe associated to a data source will query it to access column values.
Definition at line 1647 of file RDataFrame.cxx.
| ROOT::RDataFrame::RDataFrame | ( | ROOT::RDF::Experimental::RDatasetSpec | spec | ) |
Build dataframe from an RDatasetSpec object.
| [in] | spec | The dataset specification object. |
A dataset specification includes trees and file names, as well as an optional friend list and/or entry range.
See also ROOT::RDataFrame::FromSpec().
Definition at line 1671 of file RDataFrame.cxx.